生物样品全玻片成像的米勒矩阵显微镜视野扩展
IF 3
Q3 Physics and Astronomy
引用次数: 0
摘要
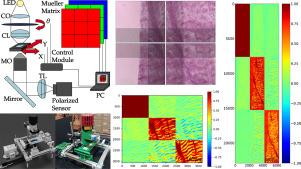
近年来,偏振光与组织相互作用的定量测量已成为生物医学诊断的有力工具。在这方面,我们实现了一种显微镜设置,该设置在成像平面中包含一个偏振传感器,以获得组织样本的给定视场(FoV)对应的Stokes参数。通过在输入中使用线性无关的偏振状态进行照明,穆勒矩阵元素也可以从样品的相同视场中检索到。为了实现全滑动成像,可以通过拼接XY位移后的多幅图像来扩展视场。我们建议在拼接算法中引入偏振特征,特别是每个像素的Mueller矩阵范数。这允许在相邻图像之间最小重叠的情况下扩展FoV,从而大大减少整个样本所需的图像总数。该方法可以显著减少全片MM成像的采集时间和数据存储要求。对组织样本整片MM检索的验证结果显示,SSIM=0.93±0.04,拼接成功率为100%,拼接成功率低至35%。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Field of view extension in Mueller matrix microscopy for whole-slide imaging of biological samples
Quantitative measures of the interaction of polarized light with tissue have been established as a powerful tool for biomedical diagnosis in recent years. In this regard, we implemented a microscopy setup that incorporates a polarized sensor in the imaging plane to obtain Stokes parameters corresponding to a given Field of View (FoV) of a tissue sample. By illuminating with linearly independent States of Polarization in the input, Mueller matrix elements can also be retrieved from the same FoV of the sample. In order to achieve whole-slide imaging the FoV can be extended by stitching multiple images taken after XY displacement. We propose introducing polarimetric features, specifically the Mueller matrix norm for each pixel, into the stitching algorithm. This allows for FoV extension with minimal overlap between neighboring images, substantially reducing the total number of images required for the entire sample. This approach can significantly reduce acquisition time and data storage requirements for whole-slide MM imaging. Validation results for the retrieval of whole-slide MM of tissue samples show and 100% stitching success from images with overlapping as low as 35%.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Results in Optics
Physics and Astronomy-Atomic and Molecular Physics, and Optics
CiteScore
2.50
自引率
0.00%
发文量
115
审稿时长
71 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


