近交系小鼠基因组串联重复序列图谱:一种遗传变异资源
IF 4.1
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
串联重复序列(TR)是人类群体遗传变异的重要来源,TR等位基因与60多种人类遗传疾病和许多生物医学性状的个体间差异有关。因此,我们利用长读测序和最先进的计算程序生成了一个包含39个近交小鼠品系的2,528,854个TRs的数据库。与人类一样,小鼠的TRs数量丰富,主要位于基因间区。然而,存在重要的物种差异:小鼠TRs不具有与人类重复扩增疾病相关的大量重复扩增,并且它们与转座因子无关。通过对40多年前确定的两种生物医学表型的分析,我们证明了这个TR数据库增强了我们表征近交系间性状差异遗传基础的能力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
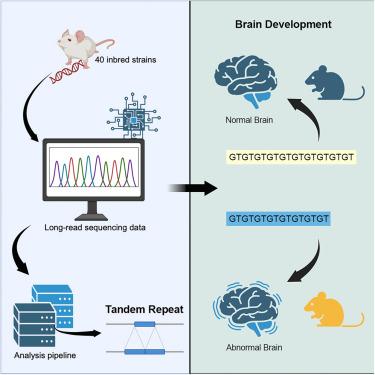
A tandem repeat atlas for the genome of inbred mouse strains: A genetic variation resource
Tandem repeats (TRs) are a significant source of genetic variation in the human population, and TR alleles are responsible for over 60 human genetic diseases and for inter-individual differences in many biomedical traits. Therefore, we utilized long-read sequencing and state of the art computational programs to produce a database with 2,528,854 TRs covering 39 inbred mouse strains. As in humans, murine TRs are abundant and were primarily located in intergenic regions. However, there were important species differences: murine TRs did not have the extensive number of repeat expansions such as those associated with human repeat expansion diseases, and they were not associated with transposable elements. We demonstrate by analysis of two biomedical phenotypes, which were identified over 40 years ago, that this TR database enhances our ability to characterize the genetic basis for trait differences among inbred strains.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


