机器学习辅助的多通道纳米酶传感器阵列用于多种农药跟踪、追踪和代谢分析。
IF 10.5
1区 生物学
Q1 BIOPHYSICS
引用次数: 0
摘要
为了实现精确的农药残留检测和代谢分析,我们创新地提出了一种机器学习辅助的多通道纳米酶传感器阵列。5种具有漆酶样和过氧化物酶样活性的cu -羧酸纳米酶对nico磺隆、2,4-二氯苯氧乙酸、毒死蜱、氯氰菊酯及其代谢物表现出明显不同的反应。在此基础上,构建了一个10通道传感器阵列。结合贝叶斯优化随机森林(BO-RF)分类模型,可同时鉴定4种农药及其代谢物。值得注意的是,4种农药的定性识别不受浓度和代谢程度的影响,具有良好的追溯能力。此外,BO-RF模型在评估农药代谢阶段方面表现出出色的预测性能。4种农药从未代谢状态到完全代谢状态的预测准确率均超过97%。进一步验证了该策略在菠菜、农业湖泊水和耕作土壤中的实用性。这项工作为农药跟踪、可追溯性和代谢分析提供了一种简单、高效和智能的方法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
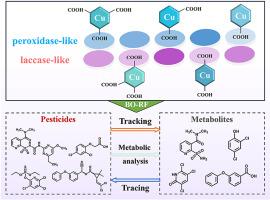
Machine learning-assisted multi-channel nanozyme sensor arrays for multiple pesticide tracking, tracing and metabolism analysis
To achieve precise pesticide residue detection and metabolic analysis, we innovatively proposed a machine learning-assisted multi-channel nanozyme sensor array. Five Cu-carboxylate nanozymes with outstanding laccase-like and peroxidase-like activities exhibited significantly distinct responses towards nicosulfuron, 2,4-dichlorophenoxyacetic acid, chlorpyrifos, cypermethrin, and their metabolites. Based on these, a 10-channel sensor array was constructed. Coupled with a Bayesian-optimized random forest (BO-RF) classification model, it enabled simultaneous identification of 4 pesticides and metabolites. Notably, qualitative recognition of 4 pesticides was not affected by variations in concentration or metabolic degree, which exhibited excellent traceback capability. Moreover, the BO-RF model showed outstanding predictive performance in assessing pesticide metabolic stages. The prediction accuracies were all exceed 97 % for 4 pesticides from unmetabolized to fully metabolized states. The practical applicability of the proposed strategy was further validated in spinach, agricultural lake water, and cultivation soil. This work offered a simple, efficient, and intelligent approach for pesticide tracking, traceability, and metabolic analysis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Biosensors and Bioelectronics
工程技术-电化学
CiteScore
20.80
自引率
7.10%
发文量
1006
审稿时长
29 days
期刊介绍:
Biosensors & Bioelectronics, along with its open access companion journal Biosensors & Bioelectronics: X, is the leading international publication in the field of biosensors and bioelectronics. It covers research, design, development, and application of biosensors, which are analytical devices incorporating biological materials with physicochemical transducers. These devices, including sensors, DNA chips, electronic noses, and lab-on-a-chip, produce digital signals proportional to specific analytes. Examples include immunosensors and enzyme-based biosensors, applied in various fields such as medicine, environmental monitoring, and food industry. The journal also focuses on molecular and supramolecular structures for enhancing device performance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


