等离子体纳米阱中核酸动力学的拉曼相关光谱单分子监测
IF 16
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
利用等离子体纳米孔中的单分子表面增强拉曼光谱(SM-SERS)对单分子进行无标记监测,可以跟踪分子动力学并深入了解其内部机制,用于催化和测序等应用。然而,不稳定等离子体热点、快速分子运动和柠檬酸盐干扰等挑战阻碍了SM-SERS数据分析和生物医学应用。在这项研究中,我们报告了一种新的SM-SERS方法,通过将单个金纳米颗粒粘附在空气中的金纳米孔中,在颗粒表面产生固定的等离子体间隙模式热点,用于连续的单分子读出和长期监测DNA扩散。DNA在热点中的无限停留时间揭示了不同DNA在单碱基分辨率下根据序列的单向和来回扩散模式以及柠檬酸盐和DNA对热点的占用。值得注意的是,发现热点的空间分辨率能够覆盖2个相邻的核碱基,DNA中的1个糖-磷酸主链和1个柠檬酸盐。通过拉曼相关光谱计算,核碱基在dna中的扩散时间根据分子结构的不同为5 ~ 22 s,而柠檬酸盐的扩散时间为0.1 ~ 7 s。我们的结果在监测生物分子动力学方面非常有希望,它们可以用于研究寡核苷酸杂交动力学,并可能为开发SM-SERS测序技术奠定基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
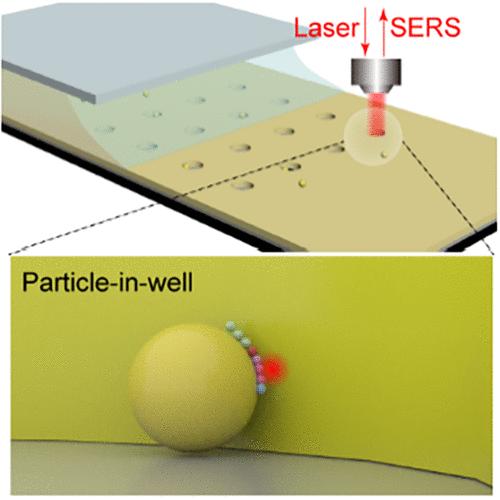
Single-Molecule Monitoring of Nucleic Acid Dynamics Using Raman Correlation Spectroscopy in Plasmonic Nanowells
Label-free monitoring of single molecules by single-molecule surface-enhanced Raman spectroscopy (SM-SERS) in plasmonic nanopores can track the molecular dynamics and gain insight into its internal mechanism for applications including catalysis and sequencing. However, challenges including unstable plasmonic hot spot, fast molecule movement, and citrate interference hinder the SM-SERS data analysis and biomedical applications. In this study, we report a new SM-SERS method by sticking a single gold nanoparticle in a gold nanowell in air to generate a fixed plasmonic gap-mode hot spot on the particle surface for continuous single-molecule readout and long-term monitoring of DNA diffusion. The unlimited resident time of the DNA in the hot spot revealed unidirectional and back-and-forth diffusion patterns of different DNAs at single-base resolution depending on their sequences as well as cooccupation of the hot spot by citrate and DNA. Significantly, the spatial resolution of the hot spot was found to be able to cover 2 neighboring nucleobases, 1 sugar–phosphate backbone in the DNA, and 1 citrate. By using Raman correlation spectroscopy, the diffusion times of nucleobases in the DNAs were calculated as 5–22 s depending on molecular structures, while those of citrate were 0.1–7 s. Our results were so promising for monitoring biomolecular dynamics that they could be used to investigate oligonucleotide hybridization kinetics and may set the basis for developing SM-SERS sequencing technologies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


