甜菜花叶病毒(BtMV)、甜菜轻度黄化病毒(BMYV)和甜菜黄化病毒(BYV)侵染本烟和甜菜的小RNA谱比较。
IF 2.7
4区 医学
Q3 VIROLOGY
引用次数: 0
摘要
植物不断受到病毒病原体的挑战,这些病原体可以限制生长并降低产量。植物先天免疫的一个关键组成部分是RNA沉默,其中病毒双链RNA (dsRNA)中间体被识别并加工成病毒衍生的小干扰RNA (vsirna)。这些vsirna指导病毒基因组的降解,从而限制感染。甜菜(Beta vulgaris subsp)是一种具有重要经济意义的作物,其中病毒黄(VY)复合体对生产构成严重威胁。在这里,我们分析和比较了天然寄主植物B. vulgaris和实验寄主植物Nicotiana benthamiana感染三种不同分类的病毒时产生的vsirna:甜菜黄病毒(BYV, clostervirus),甜菜轻度黄病毒(BMYV, Polerovirus)和甜菜花叶病毒(BtMV, Potyvirus)。小rna的高通量测序揭示了病毒和宿主物种之间不同的特征大小分布和链偏差。比较分析强调没有宿主植物特异性的vsiRNA积累模式。这种比较方法提供了vsiRNA加工的详细视图,并提供了仅从覆盖概况中不明显的新见解。在每个病毒基因组中检测到不同的vsiRNA热点,并且这些热点在宿主植物之间没有差异,这为基于RNA干扰的控制方法确定了潜在的靶点区域。这些区域的鉴定为设计合成的dsRNAs提供了基础,这些dsRNAs可以作为外源的保护性喷剂应用,这是一种新兴的非转基因策略,可以减轻VY感染,同时也促进了对甜菜和benthamiana中vsiRNA生物发生的理解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
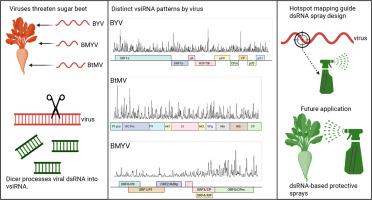
Comparative small RNA profiles of beet mosaic virus (BtMV), beet mild yellowing virus (BMYV) and beet yellows virus (BYV) infected Nicotiana benthamiana and Beta vulgaris
Plants are constantly challenged by viral pathogens that can limit growth and reduce yield. A key component of the plant innate immunity is RNA silencing, in which viral double-stranded RNA (dsRNA) intermediates are recognised and processed into virus-derived small interfering RNAs (vsiRNAs). These vsiRNAs direct the degradation of viral genomes, thereby restricting infection. Sugar beet (Beta vulgaris subsp. vulgaris) is a crop of major economic importance, where the virus yellows (VY) complex represents a serious threat to production. Here, we profiled and compared vsiRNAs generated during infection of the natural host plant B. vulgaris and the experimental host plant Nicotiana benthamiana with three taxonomically distinct viruses: beet yellows virus (BYV, Closterovirus), beet mild yellowing virus (BMYV, Polerovirus), and beet mosaic virus (BtMV, Potyvirus). High-throughput sequencing of small RNAs revealed characteristic size distributions and strand biases that differed among viruses and host species. Comparative analysis highlighted no host plant-specific pattern of vsiRNA accumulation. This comparative approach provides a detailed view of vsiRNA processing and offers novel insights that are not apparent from coverage profiles alone. Distinct vsiRNA hotspots were detected for each viral genome, and these hotspots did not differ between host plants, pinpointing potential target regions for RNA interference-based control approaches. The identification of such regions provides a basis for the design of synthetic dsRNAs that can be applied exogenously as protective sprays, an emerging, non-transgenic strategy to mitigate VY infections, while advancing understanding of vsiRNA biogenesis in sugar beet and N. benthamiana in general.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Virus research
医学-病毒学
CiteScore
9.50
自引率
2.00%
发文量
239
审稿时长
43 days
期刊介绍:
Virus Research provides a means of fast publication for original papers on fundamental research in virology. Contributions on new developments concerning virus structure, replication, pathogenesis and evolution are encouraged. These include reports describing virus morphology, the function and antigenic analysis of virus structural components, virus genome structure and expression, analysis on virus replication processes, virus evolution in connection with antiviral interventions, effects of viruses on their host cells, particularly on the immune system, and the pathogenesis of virus infections, including oncogene activation and transduction.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


