基于全基因组测序的种群规模基因分析提供了对代谢健康的见解
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
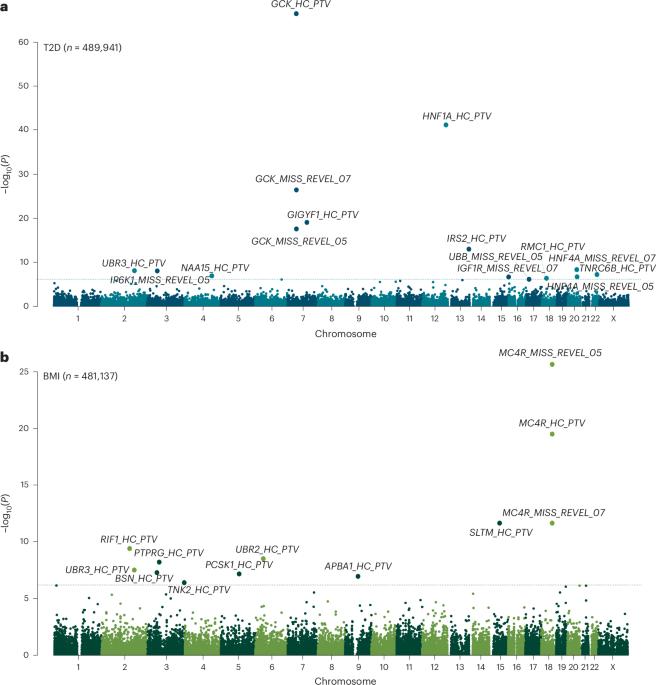
除了覆盖非编码基因组外,全基因组测序(WGS)可能比外显子组测序更好地捕获编码基因组。在这里,我们试图利用这一点,并在来自UK Biobank和All of Us研究的WGS数据(n = 708,956)中识别与代谢健康相关的新的罕见蛋白质编码变异。鉴定出的基因突出了新的生物学机制,包括DNA双链断裂修复基因RIF1中的蛋白质截短变异(PTVs),它对体重指数(2.66 kg m−2,s.e 0.43, P = 3.7 × 10−10)有实质性影响。UBR3是一个有趣的例子,ptv独立地增加了体重指数和2型糖尿病的风险。此外,IRS2中的PTVs对2型糖尿病有实质性影响(优势比6.4 (3.7-11.3),P = 9.9 × 10−14,携带者中34%的病例患病率),并且还与独立于糖尿病状态的慢性肾脏疾病相关,这表明IRS2在维持肾脏健康方面发挥重要作用。我们的研究表明,通过改进编码序列的捕获,大规模WGS为人类代谢表型提供了新的机制见解。来自UK Biobank和All of Us的全基因组测序数据分析发现了与代谢健康相关的罕见变异负担信号,包括IRS2蛋白截断变异对2型糖尿病和慢性肾脏疾病风险的影响。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Population-scale gene-based analysis of whole-genome sequencing provides insights into metabolic health
In addition to its coverage of the noncoding genome, whole-genome sequencing (WGS) may better capture the coding genome than exome sequencing. Here we sought to exploit this and identify new rare, protein-coding variants associated with metabolic health in WGS data (n = 708,956) from the UK Biobank and All of Us studies. Identified genes highlight new biological mechanisms, including protein-truncating variants (PTVs) in the DNA double-strand break repair gene RIF1 that have a substantial effect on body mass index (2.66 kg m−2, s.e. 0.43, P = 3.7 × 10−10). UBR3 is an intriguing example where PTVs independently increase body mass index and type 2 diabetes risk. Furthermore, PTVs in IRS2 have a substantial effect on type 2 diabetes (odds ratio 6.4 (3.7–11.3), P = 9.9 × 10−14, 34% case prevalence among carriers) and were also associated with chronic kidney disease independent of diabetes status, suggesting an important role for IRS2 in maintaining renal health. Our study demonstrates that large-scale WGS provides new mechanistic insights into human metabolic phenotypes through improved capture of coding sequences. Analyses of whole-genome sequencing data from UK Biobank and All of Us identify rare variant burden signals associated with metabolic health, including effects of protein-truncating variants in IRS2 on type 2 diabetes and chronic kidney disease risk.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


