使用获取后校正策略,在没有长期质量控制的情况下改善代谢组学数据的可比性
IF 6
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
代谢组学分析技术的最新进展使得生成的数据在灵敏度和稳健性方面的质量不断提高,从而为其大规模应用打开了大门。然而,由于缺乏能够在没有长期质量控制的情况下纠正分析偏差的方法,单独收集的代谢组学数据的整合目前受到限制。这一重大瓶颈阻碍了研究之间的相互比较,并限制了代谢组学在精确生物学中的影响。因此,在许多应用领域,克服这些主要挑战对于提高研究之间的互操作性以及提供更可靠和可重复的结论非常重要。在这项工作中,我们提出了一种获取后策略(PARSEC)来提高代谢组学数据的可比性,该策略包括三个步骤的工作流程,从联合提取不同研究或分析队列的原始数据,通过标准化,到基于分析质量标准的特征过滤。该工作流程应用于两个案例研究,以评估所开发的校正方法的性能,并将其与经典的局部估计散点图平滑(黄土)方法进行比较。PARSEC策略允许减少组间变异性,并产生更均匀的样本分布。此外,结果表明,在这两个案例研究中,数据的可比性有所改善,使得最初被不需要的变异性来源掩盖的生物信息比黄土方法更清楚地揭示出来。提出的采集后校正策略结合了批量标准化和混合建模,增强了代谢组学研究的数据可比性和可扩展性。通过处理批和组效应,这种方法最大限度地减少了分析条件的影响,同时保持了生物可变性。因此,它为在没有共同的长期质量控制样本的情况下协调不同研究或队列的数据集提供了一个有价值的工具。本文章由计算机程序翻译,如有差异,请以英文原文为准。
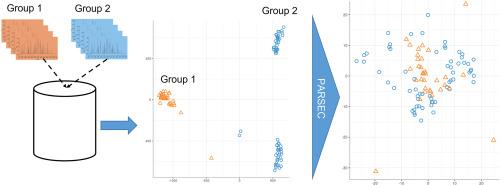
Improving metabolomics data comparability without long term quality controls using a post-acquisition correction strategy
Background
Recent advances in analytical techniques for metabolomics allowed generating data of increasing quality in terms of sensitivity and robustness, thus opening the door to its large-scale application. However, the integration of separately collected metabolomic data is currently limited by the lack of methods able to correct for the analytical bias without long-term quality controls. This significant bottleneck prevents inter-comparisons across studies and limits metabolomics impact in precision biology. Overcoming these major challenges is therefore of great importance in many application fields to improve interoperability across studies and offer more reliable and reproducible conclusions.Results
In this work, we propose a post-acquisition strategy (PARSEC) to improve metabolomics data comparability that consists in a three-step workflow starting from the combined extraction of raw data from the different studies or cohorts analyzed, through standardization, to the filtering of features based on analytical quality criteria. This workflow was applied to two case studies to evaluate the performance of the developed correction approach and to compare it with the classically used locally estimated scatterplot smoothing (LOESS) method. The PARSEC strategy allowed reducing the inter-group variability, and producing a more homogeneous sample distribution. In addition, results showed an improvement in the comparability of the data in both case studies, allowing biological information initially masked by unwanted sources of variability to be revealed more clearly than with the LOESS method.Significance
The proposed post-acquisition correction strategy, which combines batch-wise standardization and mixed modeling, enhances data comparability and scalability for metabolomics studies. By addressing both batch and group effects, this approach minimizes the influence of analytical conditions while preserving biological variability. Therefore, it offers a valuable tool for harmonizing datasets across different studies or cohorts without common long-term quality control samples.求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


