利用Nimbus对多路成像数据中的细胞表达进行自动分类
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
多路成像提供了一种强大的方法来表征健康和疾病组织的空间地形。为了分析这些数据,必须列举每个细胞中存在的特定标记组合,以实现准确的表型,这一过程通常依赖于无监督聚类。我们构建了Pan-Multiplex (Pan-M)数据集,其中包含15种不同细胞类型的1.97亿个不同的标记表达注释。我们使用Pan-M创建了Nimbus,这是一个深度学习模型,用于从多路复用图像数据中预测标记的积极性。Nimbus是一种预先训练的模型,它使用基础图像对来自不同组织的不同细胞类型的单个细胞的标记表达进行阳性或阴性分类,使用不同的显微镜平台获得,无需任何再训练。我们证明了Nimbus预测捕获了Pan-M中存在的全部标记多样性的潜在染色模式,并且Nimbus匹配或超过了以前必须在每个数据集上重新训练的方法的准确性。然后,我们展示了如何将Nimbus预测与下游聚类算法集成在一起,以稳健地识别图像数据中的细胞亚型。我们已经开源了Nimbus和Pan-M,使社区可以在https://github.com/angelolab/Nimbus-Inference上使用。Nimbus是一种深度学习模型,它使用大型多路成像数据集来预测单个细胞中标记物阳性的可能性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
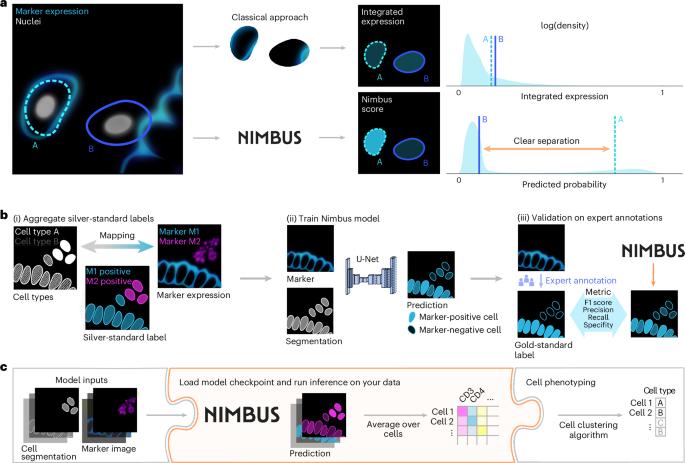
Automated classification of cellular expression in multiplexed imaging data with Nimbus
Multiplexed imaging offers a powerful approach to characterize the spatial topography of tissues in both health and disease. To analyze such data, the specific combination of markers that are present in each cell must be enumerated to enable accurate phenotyping, a process that often relies on unsupervised clustering. We constructed the Pan-Multiplex (Pan-M) dataset containing 197 million distinct annotations of marker expression across 15 different cell types. We used Pan-M to create Nimbus, a deep learning model to predict marker positivity from multiplexed image data. Nimbus is a pretrained model that uses the underlying images to classify marker expression of individual cells as positive or negative across distinct cell types, from different tissues, acquired using different microscope platforms, without requiring any retraining. We demonstrate that Nimbus predictions capture the underlying staining patterns of the full diversity of markers present in Pan-M, and that Nimbus matches or exceeds the accuracy of previous approaches that must be retrained on each dataset. We then show how Nimbus predictions can be integrated with downstream clustering algorithms to robustly identify cell subtypes in image data. We have open-sourced Nimbus and Pan-M to enable community use at https://github.com/angelolab/Nimbus-Inference . Nimbus, a deep learning model, uses a large multiplexed imaging dataset to predict the likelihood of marker positivity in single cells.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


