完整的线粒体和叶绿体基因组揭示了基因转移和RNA编辑事件
IF 6.2
1区 农林科学
Q1 AGRICULTURAL ENGINEERING
引用次数: 0
摘要
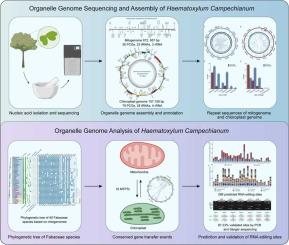
骆家辉(Haematoxylum campechianum L.)是一种具有多种药用价值的经济树种。然而,原木的细胞器基因组尚未被阐明,从而阻碍了对原木亚科的系统发育研究和比较分析,也限制了对其生物活性代谢物生物合成途径的遗传研究。在本研究中,我们成功地组装和注释了campechianum的线粒体基因组(有丝分裂基因组)和叶绿体基因组。结果表明,原木有丝分裂基因组全长672,957 bp,包括36个蛋白质编码基因(PCGs)、23个trna和3个rnas。相比之下,原木叶绿体基因组长度为157,156 bp,包含79个PCGs, 28个tRNA基因和4个rRNA基因。此外,系统发育重建揭示了线粒体突变率加速导致有丝分裂基因组树和叶绿体树之间的拓扑不一致。此外,鉴定了16个线粒体质体DNA片段,证明了细胞器间基因转移,以及286个预测的RNA编辑位点,验证率为87.23 %。本研究为杉木亚科提供了宝贵的细胞器基因组资源,阐明了杉木的系统发育关系,为探讨杉木的进化提供了必要的资源。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Complete mitochondrial and chloroplast genomes of Haematoxylum campechianum L. reveal insights into gene transfer and RNA editing events
Logwood (Haematoxylum campechianum L.) is an economically valuable tree species with diverse medicinal properties. However, the organelle genomes of logwood have not yet been elucidated, thereby hindering phylogenetic studies and comparative analyses within the Caesalpinioideae subfamily as well as limiting genetic investigations of its bioactive metabolite biosynthesis pathways. In this study, the mitochondrial genome (mitogenome) and chloroplast genome of H. campechianum were assembled and annotated successfully. The results demonstrated that the logwood mitogenome spanned 672,957 bp, including 36 protein-coding genes (PCGs), 23 tRNAs, and 3 rRNAs. In comparison, the chloroplast genome of logwood was 157,156 bp long and contained 79 PCGs, 28 tRNA genes, and 4 rRNA genes. In addition, phylogenetic reconstruction revealed topological discordance between mitogenome- and chloroplast-based trees, attributable to mitochondrial mutation rate acceleration. Furthermore, 16 mitochondrial plastid DNA segments were identified, evidencing inter-organellar gene transfer, along with 286 predicted RNA editing sites showing the 87.23 % validation rate. Taken together, this study provided the valuable organelle genome resources for the Caesalpinioideae subfamily, elucidated the phylogenetic relationships, and supplied essential resources for probing the evolution of logwood.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Industrial Crops and Products
农林科学-农业工程
CiteScore
9.50
自引率
8.50%
发文量
1518
审稿时长
43 days
期刊介绍:
Industrial Crops and Products is an International Journal publishing academic and industrial research on industrial (defined as non-food/non-feed) crops and products. Papers concern both crop-oriented and bio-based materials from crops-oriented research, and should be of interest to an international audience, hypothesis driven, and where comparisons are made statistics performed.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


