m7G-PIP:以m7g为中心的rna -蛋白相互作用组的空间分辨活细胞定位
IF 3.7
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
m⁷G修饰调节mRNA生命周期并驱动癌症和病毒感染的进展。然而,目前的检测方法依赖于抗体和破坏天然细胞结构,使m⁷G的活细胞原位定位成为一个未满足的挑战。为了解决这个问题,我们开发了m⁷G-Proximity Interactome Profiling (m⁷G-PIP)。这种无抗体技术使用接近标记来接近结合m⁷G位点的生物素化生物分子,放大信号,用于活细胞中rna -蛋白相互作用组的纳米级分辨率(< 20nm)制图。将该技术应用于单纯疱疹病毒-1 (HSV-1),我们揭示了感染期间以m⁷G为中心的动态网络,并鉴定了病毒转录物上的内部m⁷G修饰——这是在HSV-1上的第一个此类证据。m⁷G-PIP的模块化设计很容易适用于用结合蛋白分析其他RNA修饰。我们的工作建立了一个表转录组调控的时空框架,并为开发靶向RNA修饰途径的抗病毒药物提供了一个多功能平台。本文章由计算机程序翻译,如有差异,请以英文原文为准。
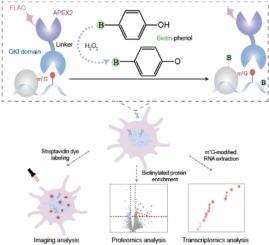
m7G-PIP: Spatially resolved live-cell mapping of m7G-centric RNA-protein interactomes
m⁷G modification regulates mRNA life cycle and drives progression of cancers and viral infections. Current detection methods, however, rely on antibodies and disrupt native cellular architecture, making live-cell in situ mapping of m⁷G an unmet challenge. To address this, we develop m⁷G-Proximity Interactome Profiling (m⁷G-PIP). This antibody-free technology uses proximity labeling to biotinylate biomolecules proximal to bound m⁷G sites, amplifying signals for nanoscale-resolution (< 20 nm) mapping of RNA-protein interactomes in live cells. Applying this technology to herpes simplex virus-1 (HSV-1), we reveal dynamic m⁷G-centered networks during infection and identify internal m⁷G modifications on viral transcripts—the first such evidence on HSV-1. The modular design of m⁷G-PIP is readily adaptable to profile other RNA modifications with binding proteins. Our work establishes a spatiotemporal framework for epitranscriptome regulation and provides a versatile platform for developing antivirals targeting RNA modification pathways.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Sensors and Actuators B: Chemical
工程技术-电化学
CiteScore
14.60
自引率
11.90%
发文量
1776
审稿时长
3.2 months
期刊介绍:
Sensors & Actuators, B: Chemical is an international journal focused on the research and development of chemical transducers. It covers chemical sensors and biosensors, chemical actuators, and analytical microsystems. The journal is interdisciplinary, aiming to publish original works showcasing substantial advancements beyond the current state of the art in these fields, with practical applicability to solving meaningful analytical problems. Review articles are accepted by invitation from an Editor of the journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


