所有在一次RNA折叠与三维基序预测框架的进化信息。
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
在环状区域中发现了大量循环的短三维(3D)元素,这些元素涉及非沃森-克里克相互作用,有助于将规范双螺旋排列成三级结构。在这里,我们提出了cofold - r3d,这是一种概率语法,可以预测这些RNA 3D基序(也称为模块)与RNA二级结构在序列或排列上的联合。cofold - r3d利用存在于RNA序列中的进化信息,通过共变可靠地识别规范螺旋(包括假结)。在这里,我们进一步介绍了R3D语法,它也利用螺旋共变来限制大多数非共变RNA 3D基序的定位。我们的方法对50多个已知RNA基序(“一切”)的几乎详尽的列表进行预测。图案可以出现在任何非螺旋环区域(包括三向,四向和更高的连接)(“无处不在”)。所有的结构基元以及规范的螺旋都被安排在一个单一的结构中,由一个联合概率语法(All -at-once)预测。我们的研究结果表明,cofold - r3d是预测RNA 3D结构中存在的全残基相互作用的有效替代方法。cofold - r3d是一种快速且易于定制的新基序发现方法,作为全原子结构预测的深度学习方法以及指导RNA设计作为治疗性小分子药物靶点的强大输入,显示出有希望的价值。本文章由计算机程序翻译,如有差异,请以英文原文为准。
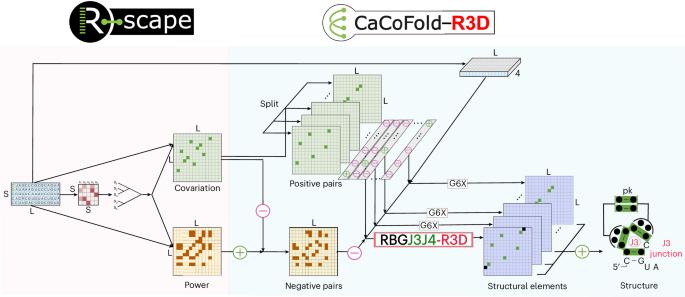
All-at-once RNA folding with 3D motif prediction framed by evolutionary information
Structural RNAs exhibit a vast array of recurrent short three-dimensional (3D) elements found in loop regions involving non-Watson–Crick interactions that help arrange canonical double helices into tertiary structures. Here we present CaCoFold-R3D, a probabilistic grammar that predicts these RNA 3D motifs (also termed modules) jointly with RNA secondary structure over a sequence or alignment. CaCoFold-R3D uses evolutionary information present in an RNA alignment to reliably identify canonical helices (including pseudoknots) by covariation. Here we further introduce the R3D grammars, which also exploit helix covariation that constrains the positioning of the mostly noncovarying RNA 3D motifs. Our method runs predictions over an almost-exhaustive list of over 50 known RNA motifs (‘everything’). Motifs can appear in any nonhelical loop region (including three-way, four-way and higher junctions) (‘everywhere’). All structural motifs as well as the canonical helices are arranged into one single structure predicted by one single joint probabilistic grammar (‘all-at-once’). Our results demonstrate that CaCoFold-R3D is a valid alternative for predicting the all-residue interactions present in a RNA 3D structure. CaCoFold-R3D is fast and easily customizable for novel motif discovery and shows promising value both as a strong input for deep learning approaches to all-atom structure prediction as well as toward guiding RNA design as drug targets for therapeutic small molecules. CaCoFold-R3D is a probabilistic model that simultaneously predicts the RNA 3D motifs jointly with the secondary structure in a structural RNA using evolutionary information.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


