鲜肉和肉类制品中耐药恩培多杆菌的鉴定
IF 7
2区 农林科学
Q1 FOOD SCIENCE & TECHNOLOGY
引用次数: 0
摘要
Empedobacter已被确定为一种机会性病原体,经常对多种抗生素产生耐药性,包括一些被称为最后手段的抗生素。本研究描述了从零售鲜肉和肉类制品中分离出的耐碳青霉烯Empedobacter的表型和基因型特征。采用微量肉汤稀释法测定62株菌株对15种常用抗生素的敏感性。此外,对其中24株菌株进行了全基因组测序(WGS),以确定它们的分类,并鉴定抗微生物药物耐药性基因(ARGs),以及它们的染色体或质粒传播位置。经常检测到对美罗培南、环丙沙星、阿米卡星、庆大霉素、氯霉素、四环素和/或粘菌素的耐药性,尽管某些抗生素没有检测到断点,但仍有61.3%的恩培多杆菌菌株被列为多重耐药(MDR)。WGS结果显示,所有假恩培杆菌分离株和单个罗非鱼恩培杆菌分离株中均存在blaEBR-1基因,1株假恩培杆菌分离株中存在染色体e(D)基因,8株假恩培杆菌分离株中存在tet(X2)基因,其中7株在质粒中。这些发现强调需要进一步研究,以确定被忽视的非eskape细菌(如Empedobacter)在肉类生产系统中抗菌素耐药性传播中的作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
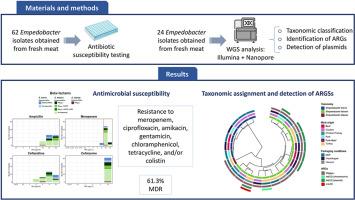
Characterization of antimicrobial resistant Empedobacter from fresh meat and meat preparations
Empedobacter has been identified as an opportunistic pathogen that frequently exhibits resistance to multiple antibiotics, including some of those known as of last-resort. This study describes the phenotypic and genotypic characterization of carbapenem-resistant Empedobacter isolates obtained from retail fresh meat and meat preparations. The antimicrobial susceptibility of 62 isolates to 15 common antibiotics was assessed using the broth microdilution method. Additionally, whole genome sequencing (WGS) was performed on 24 of these isolates to determine their taxonomic classification and to identify antimicrobial resistance genes (ARGs), as well as their chromosomal or plasmid-borne location. Resistance to meropenem, ciprofloxacin, amikacin, gentamicin, chloramphenicol, tetracycline, and/or colistin was frequently detected, with 61.3 % of the Empedobacter strains being classified as multi-drug resistant (MDR) despite the absence of breakpoints for some of the antibiotics tested. WGS revealed the presence of blaEBR-1 genes in all Empedobacter falsenii isolates and the single Empedobacter tilapiae isolate, of a chromosomic ere(D) gene in one E. falsenii isolate, and of tet(X2) genes in eight E. falsenii isolates, seven of them harboured in plasmids.
These findings underscore the need for further research to determine the role of neglected non-ESKAPE bacteria, such as Empedobacter, in the spread of antimicrobial resistance in meat production systems.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Current Research in Food Science
Agricultural and Biological Sciences-Food Science
CiteScore
7.40
自引率
3.20%
发文量
232
审稿时长
84 days
期刊介绍:
Current Research in Food Science is an international peer-reviewed journal dedicated to advancing the breadth of knowledge in the field of food science. It serves as a platform for publishing original research articles and short communications that encompass a wide array of topics, including food chemistry, physics, microbiology, nutrition, nutraceuticals, process and package engineering, materials science, food sustainability, and food security. By covering these diverse areas, the journal aims to provide a comprehensive source of the latest scientific findings and technological advancements that are shaping the future of the food industry. The journal's scope is designed to address the multidisciplinary nature of food science, reflecting its commitment to promoting innovation and ensuring the safety and quality of the food supply.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


