通过靶向乳酸菌接种减轻青贮饲料中四环素抗性基因。
IF 9
1区 环境科学与生态学
Q1 AGRICULTURAL ENGINEERING
引用次数: 0
摘要
抗生素耐药基因(ARGs)在青贮生态系统中的传播对生态稳定和公共卫生安全构成了重大挑战。本研究以青贮饲料中最常见的四环素抗性基因(TRGs)为研究对象,采用有针对性的乳酸菌(LAB)接种剂选择策略。从226株分离的乳酸菌中,4株候选菌株(LP1-3:植物乳酸菌;LC1:副干酪乳杆菌)表现出优异的生长动力学(24 h内OD600 bbb1.6)和快速酸化能力(pH 0.4, p本文章由计算机程序翻译,如有差异,请以英文原文为准。
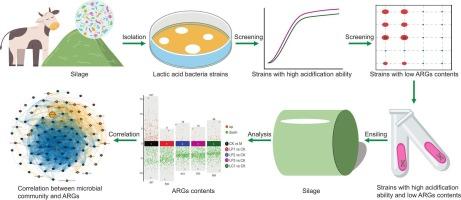
Mitigation of tetracycline resistance genes in silage through targeted lactic acid bacteria inoculation
The dissemination of antibiotic resistance genes (ARGs) in silage ecosystems poses a critical challenge to ecological stability and public health security. This investigation focuses on tetracycline resistance genes (TRGs), the most prevalent subtype of ARGs in silage, employing a targeted selection strategy for lactic acid bacteria (LAB) inoculants. From 226 isolated LAB strains, four candidates (LP1-3: Lactiplantibacillus plantarum; LC1: Lacticaseibacillus paracasei) demonstrating superior growth kinetics (OD600 > 1.6 within 24 h) and rapid acidification capacity (pH < 3.9 within 24 h) were selected. Strains LP3 and LC1 exhibited minimal intrinsic TRGs content. These four strains reduced (p < 0.001) pH, ammonia-N concentration, and coliform bacterial counts of stylo silage. Metagenomic analysis revealed that strains LP1-3 promoted Lactiplantibacillus dominance (0.709–0.975 vs. 0.379–0.509 in the control), while LC1 enhanced Lacticaseibacillus abundance (0.449–0.612 vs. 0.002–0.013 in the control). Ensiling process downregulated 367 and upregulated 227 ARGs. Inoculation with the four LAB strains further enhanced the suppression of ARGs. Among the top 30 TRGs, 22 were reduced by strains LP1-3 and 20 by LC1. Quantitative PCR results showed that strains LP1-3 decreased (p < 0.05) the contents of tetA and tetG during 30 days fermentation. Among the TRGs increased, tetA(60), tetB(58), tet(T) were positively correlated with Lactiplantibacillus spp., tetA(58), tetB(60), tetA(46), tetB(46), tet(43) were significantly correlated with Lacticaseibacillus spp. (R > 0.4, p < 0.001). In conclusion, the fermentation process induced substantial ARGs profile modifications, LAB-mediated microbiome engineering enables TRGs suppression, providing a scientific foundation for precision silage management strategies targeting antimicrobial resistance mitigation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Bioresource Technology
工程技术-能源与燃料
CiteScore
20.80
自引率
19.30%
发文量
2013
审稿时长
12 days
期刊介绍:
Bioresource Technology publishes original articles, review articles, case studies, and short communications covering the fundamentals, applications, and management of bioresource technology. The journal seeks to advance and disseminate knowledge across various areas related to biomass, biological waste treatment, bioenergy, biotransformations, bioresource systems analysis, and associated conversion or production technologies.
Topics include:
• Biofuels: liquid and gaseous biofuels production, modeling and economics
• Bioprocesses and bioproducts: biocatalysis and fermentations
• Biomass and feedstocks utilization: bioconversion of agro-industrial residues
• Environmental protection: biological waste treatment
• Thermochemical conversion of biomass: combustion, pyrolysis, gasification, catalysis.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


