光催化接近标记与交联耦合用于活细胞中溶酶体蛋白质组的解码
IF 6
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
溶酶体是在多种生物分子的降解和再循环中起重要作用的重要细胞器。为了充分了解它们的分子机制,对活细胞中溶酶体蛋白质组进行高时空鉴定是至关重要的。虽然光催化接近标记是分析亚细胞蛋白质组的有力工具,但它经常与低标记效率作斗争,特别是对于低丰度的蛋白质。为了克服这一问题,我们引入了一种将光催化接近标记与交联相结合的新策略。以溶酶体为靶点的光敏剂(MP-AcDBF)催化溶酶体内近端蛋白与可富集的亲核底物之间基于单线态氧的共价反应。然后,交联剂允许难以标记的蛋白质(低丰度或瞬时定位的蛋白质)连接到预标记的蛋白质,从而扩大蛋白质鉴定的范围,同时通过富集标记的处理保留溶酶体的特异性。使用交联辅助光催化接近标记(CLAPL)策略,我们成功地在活HeLa细胞中鉴定了238种溶酶体注释的蛋白,包括腔体,跨膜和膜相关蛋白,与没有交联的方法相比,能够相对增加(238对197)。此外,CLAPL进一步应用于动态溶酶体蛋白质组图谱,揭示了应激诱导的代谢改变。开发的策略通过高精度集成平台增强了溶酶体蛋白质组学的覆盖范围,允许系统地鉴定溶酶体内的膜相关蛋白和腔内蛋白,从而为亚细胞生物学提供机制见解,并开辟细胞研究的新领域。本文章由计算机程序翻译,如有差异,请以英文原文为准。
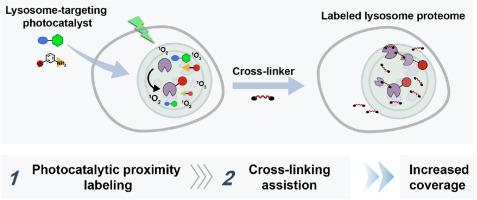
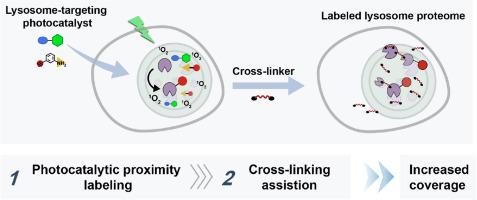
Photocatalytic proximity labeling coupled with cross-linking for deciphering of the lysosome proteome in living cells
Background
Lysosomes are essential organelles that play vital roles in the degradation and recycling of a variety of biomolecules. To fully understand their molecular mechanisms, high spatiotemporal identification of the lysosome proteome in living cells is critical. Although photocatalytic proximity labeling is a powerful tool for profiling subcellular proteome, it often struggles with low labeling efficiency, particularly for low-abundance proteins.
Results
To overcome this, we introduced a novel strategy that integrates photocatalytic proximity labeling with cross-linking. The lysosome-targeting photosensitizer (MP-AcDBF) catalyzed the covalent reaction between proximal proteins and enrichable nucleophilic substrate within lysosome based on singlet oxygen. Then the cross-linker allowed difficult-to-label proteins (low-abundance or transiently localized proteins) to be linked to pre-labeled ones, thereby expanding the scope of protein identification while preserving lysosomal specificity via enrichment of labeled handles. Using the cross-linking-assisted photocatalytic proximity labeling (CLAPL) strategy, we successfully identified 238 lysosome-annotated proteins in living HeLa cells, covering luminal, transmembrane and membrane-associated proteins, enabling a relative increase compared to method without cross-linking (238 vs 197). Additionally, CLAPL was further applied to map dynamic lysosomal proteome and revealed stress-induced metabolic alterations.
Significance
The developed strategy enhanced lysosomal proteomic coverage via a high-precision integration platform, allowing systematic identification of both membrane-associated and luminal proteins within lysosome, thereby providing mechanistic insights into subcellular biology and unlocking new frontiers in cellular research.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


