发现隐藏的蛋白质修饰与天然自上而下的质谱。
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
蛋白质修饰通过调节生物分子相互作用驱动动态细胞过程,然而在其天然结构背景下捕获这些修饰仍然是一个重大挑战。原生自顶向下质谱法有望保留修饰和相互作用之间的关键联系。然而,目前的方法往往无法检测到未表征或低丰度的修饰,从而限制了对蛋白质种类多样性的了解。为了解决这一差距,我们引入了精确和准确的原生蛋白质形态识别(precisION),这是一个交互式的端到端软件包,利用强大的、数据驱动的片段级开放搜索来检测、定位和量化完整蛋白质复合物中的“隐藏”修饰。将precisION应用于四个治疗相关靶标- pde6, ACE2,骨桥蛋白(SPP1)和GABA转运蛋白(GAT1)-我们发现了未记录的磷酸化,糖基化和脂化,并在GAT1的电子冷冻显微镜图中解决了以前无法解释的密度。作为一个开源软件包,precisION提供了一种直观的方法来解释复杂的蛋白质碎片数据。该工具将使社区能够释放天然自上而下质谱法的潜力,推进综合结构生物学,分子病理学和药物开发。本文章由计算机程序翻译,如有差异,请以英文原文为准。
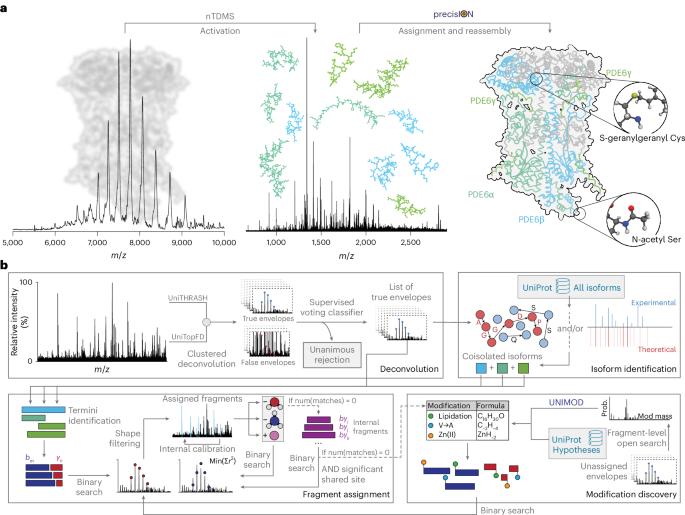
Uncovering hidden protein modifications with native top-down mass spectrometry
Protein modifications drive dynamic cellular processes by modulating biomolecular interactions, yet capturing these modifications within their native structural context remains a significant challenge. Native top-down mass spectrometry promises to preserve the critical link between modifications and interactions. However, current methods often fail to detect uncharacterized or low-abundance modifications, limiting insights into proteoform diversity. To address this gap, we introduce precise and accurate Identification Of Native proteoforms (precisION), an interactive end-to-end software package that leverages a robust, data-driven fragment-level open search to detect, localize and quantify ‘hidden’ modifications within intact protein complexes. Applying precisION to four therapeutically relevant targets—PDE6, ACE2, osteopontin (SPP1) and a GABA transporter (GAT1)—we discover undocumented phosphorylation, glycosylation and lipidation, and resolve previously uninterpretable density in an electron cryo-microscopy map of GAT1. As an open-source software package, precisION offers an intuitive means for interpreting complex protein fragmentation data. This tool will empower the community to unlock the potential of native top-down mass spectrometry, advancing integrative structural biology, molecular pathology and drug development. precisION discovers, localizes and quantifies protein modifications within complex proteoform assemblies through data-driven analysis of native top-down mass spectra.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


