结核分枝杆菌转录调控的进化与传播和耐药性增加有关
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
结核分枝杆菌(Mtb)已与人类共同进化了数千年,其特点是在毒力、传播性和疾病表型方面存在差异。为了确定细菌对表型多样性的贡献,我们开发了新的RNA测序(RNA-seq)和系统基因组学工具来捕获数百个结核分枝杆菌分离物转录组,将转录和遗传变异联系起来,并发现变异与流行病学特征之间的关联。在274株结核分枝杆菌临床分离株中,我们发现了毒力基因表达的意想不到的多样性,我们将其与已知和未知的调节因子联系起来。令人惊讶的是,我们发现许多分离株携带与EsxA (Esat6)和EsxB (Cfp10)表达降低相关的变异,这是毒力效应物,优势T细胞抗原和免疫诊断靶点。在55,000株分离株中,这些变异与增加的传播性有关,特别是在耐药结核分枝杆菌菌株中。我们的数据表明,结核分枝杆菌毒力基因的表达正在随着药物相关压力而进化,这引起了人们对在免疫诊断和下一代疫苗中使用这些靶点的担忧。本文章由计算机程序翻译,如有差异,请以英文原文为准。
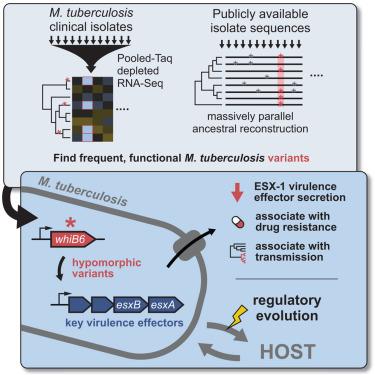
Evolution of Mycobacterium tuberculosis transcription regulation is associated with increased transmission and drug resistance
Mycobacterium tuberculosis (Mtb) has co-evolved with humans for thousands of years and is characterized by variation in virulence, transmissibility, and disease phenotypes. To identify bacterial contributors to phenotypic diversity, we developed new RNA sequencing (RNA-seq) and phylogenomic tools to capture hundreds of Mtb isolate transcriptomes, link transcriptional and genetic variation, and find associations between variants and epidemiologic traits. Across 274 Mtb clinical isolates, we uncovered unexpected diversity in virulence gene expression, which we linked to known and unknown regulators. Surprisingly, we found that many isolates harbor variants associated with decreased expression of EsxA (Esat6) and EsxB (Cfp10), which are virulence effectors, dominant T cell antigens, and immunodiagnostic targets. Across >55,000 isolates, these variants associate with increased transmissibility, especially in drug-resistant Mtb strains. Our data suggest expression of Mtb virulence genes is evolving in response to drug-linked pressure, raising concerns about use of these targets in immunodiagnostics and next-generation vaccines.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


