功能性RNA分裂推动了转座子V型CRISPR-Cas系统的进化出现
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
转座子编码的TnpB核酸酶通过多个独立的驯化事件产生V型CRISPR-Cas12效应物。这些系统使用不同的RNA分子作为DNA靶向的指南:TnpB的转座子衍生的右端RNA (RNAs或omega RNAs)和V型CRISPR- cas系统的CRISPR RNA。然而,连接转座子活性和CRISPR免疫的分子机制尚不清楚。我们鉴定了来自不同IS605-或IS607-TnpB谱系的TranCs(转座子crispr中间产物)。TranCs利用CRISPR rna和RNAs来指导DNA切割。Lawsonibacter sp.的LaTranC的低温电镜(cryo-EM)结构与ISDra2 TnpB复合物非常相似;然而,与单分子RNA不同,lanranc引导RNA在功能上分为tracrRNA和crRNA。ISDra2 TnpB的工程RNA分裂使其具有CRISPR阵列的活性。这些发现表明,功能性RNA分裂是驱动转座子出现多种V型CRISPR-Cas系统的主要分子事件。本文章由计算机程序翻译,如有差异,请以英文原文为准。
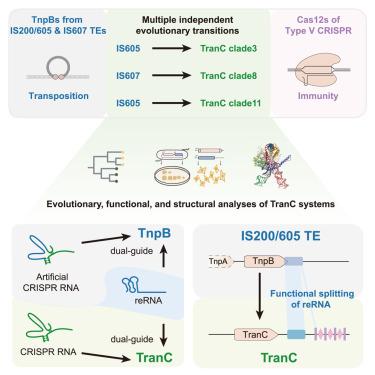
Functional RNA splitting drove the evolutionary emergence of type V CRISPR-Cas systems from transposons
Transposon-encoded TnpB nucleases gave rise to type V CRISPR-Cas12 effectors through multiple independent domestication events. These systems use different RNA molecules as guides for DNA targeting: transposon-derived right-end RNAs (reRNAs or omega RNAs) for TnpB and CRISPR RNAs for type V CRISPR-Cas systems. However, the molecular mechanisms bridging transposon activity and CRISPR immunity remain unclear. We identify TranCs (transposon-CRISPR intermediates) derived from distinct IS605- or IS607-TnpB lineages. TranCs utilize both CRISPR RNAs and reRNAs to direct DNA cleavage. The cryoelectron microscopy (cryo-EM) structure of LaTranC from Lawsonibacter sp. closely resembles that of the ISDra2 TnpB complex; however, unlike a single-molecule reRNA, the LaTranC guide RNA is functionally split into a tracrRNA and crRNA. An engineered RNA split of ISDra2 TnpB enabled activity with a CRISPR array. These findings indicate that functional RNA splitting was the primary molecular event driving the emergence of diverse type V CRISPR-Cas systems from transposons.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


