基于稳健和低成本靶组织捕获的全长空间转录组策略
IF 10.5
1区 生物学
Q1 BIOPHYSICS
引用次数: 0
摘要
Smart-seq2是一种广泛用于构建全长mRNA转录组文库的方法,可以与靶组织捕获技术(如激光捕获显微解剖(LCM))相结合,以实现强大的空间转录组分析。然而,Smart-seq2和LCM都受到高成本和复杂操作的限制,阻碍了它们的广泛采用。在这里,我们报告了一种基于强大且具有成本效益的组织捕获技术的全长空间转录组策略,称为MSN-seq。MSN-seq集成了基于微针的采样与改进的Smart-seq2协议。该方法捕获多细胞组织点,不仅优化了Smart-seq2以提高性能,而且利用可重复使用的钢针进一步降低成本并提高转录组捕获效率。除了简化实验程序外,MSN-seq通过实现空间转录组分析而不需要专业技能或昂贵的仪器,从而增加了可访问性。我们将MSN-seq应用于帕金森病和视网膜变性模型的大脑和视网膜组织,证明其在捕获空间转录组数据方面的有效性。此外,将MSN-seq应用于U251胶质母细胞瘤模型小鼠的冷冻切片,揭示了早期侵袭机制和免疫细胞亚群的空间限制性富集。总的来说,MSN-seq为获取空间转录组数据提供了一种低成本、易于使用和高效的策略。本文章由计算机程序翻译,如有差异,请以英文原文为准。
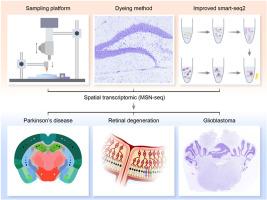
Full-length spatial transcriptome strategy based on robust and low-cost target tissue capture
Smart-seq2, a widely used method for constructing full-length mRNA transcriptome libraries, can be combined with target tissue capture techniques such as Laser Capture Microdissection (LCM) to achieve robust spatial transcriptomic profiling. However, both Smart-seq2 and LCM are limited by high costs and complex operation, hindering their broader adoption. Here, we report a full-length spatial transcriptome strategy based on a robust and cost-effective tissue capture technique, termed MSN-seq. MSN-seq integrates microneedle-based sampling with a modified Smart-seq2 protocol. This method captures multi-cellular tissue spots and not only optimises Smart-seq2 for enhanced performance but also leverages reusable steel needles to further reduce costs and improve transcriptomic capture efficiency. In addition to simplifying experimental procedures, MSN-seq increases accessibility by enabling spatial transcriptomic analysis without requiring specialised skills or expensive instrumentation. We applied MSN-seq to brain and retinal tissues from models of Parkinson's disease and retinal degeneration, demonstrating its effectiveness in capturing spatial transcriptomic data. Furthermore, application of MSN-seq to frozen sections from U251 glioblastoma model mice revealed early invasive mechanisms and spatially restricted enrichment of immune cell subsets. Overall, MSN-seq provides a low-cost, easy-to-use, and highly efficient strategy for acquiring spatial transcriptome data.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Biosensors and Bioelectronics
工程技术-电化学
CiteScore
20.80
自引率
7.10%
发文量
1006
审稿时长
29 days
期刊介绍:
Biosensors & Bioelectronics, along with its open access companion journal Biosensors & Bioelectronics: X, is the leading international publication in the field of biosensors and bioelectronics. It covers research, design, development, and application of biosensors, which are analytical devices incorporating biological materials with physicochemical transducers. These devices, including sensors, DNA chips, electronic noses, and lab-on-a-chip, produce digital signals proportional to specific analytes. Examples include immunosensors and enzyme-based biosensors, applied in various fields such as medicine, environmental monitoring, and food industry. The journal also focuses on molecular and supramolecular structures for enhancing device performance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


