DNA甲基化和组蛋白修饰的单细胞多组学检测重建表观基因组维持的动力学。
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
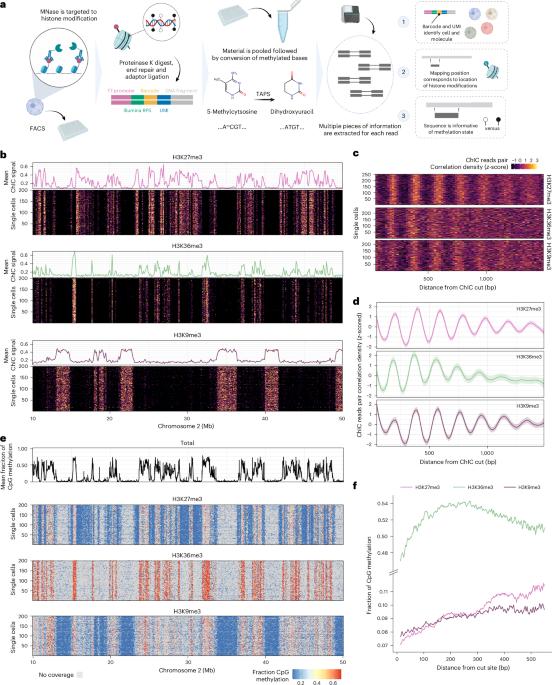
DNA甲基化和组蛋白修饰编码表观遗传信息。最近,在单细胞分辨率测量标记方面取得了重大进展;然而,缺乏一种同时检测的方法,阻碍了对它们相互作用的研究。在这里,为了弥补这个差距,我们开发了scEpi2-seq。我们的技术在单细胞和单分子水平上提供组蛋白修饰和DNA甲基化的读数。在具有FUCCI细胞周期报告系统的细胞系中的应用揭示了DNA甲基化维持如何受到局部染色质环境的影响。此外,对小鼠肠道中H3K27me3和DNA甲基化的分析可以深入了解细胞类型规范过程中的表观遗传相互作用。除了H3K27me3调控外,差异甲基化区域还表现出独立的细胞型调控,这进一步表明CpG甲基化在兼性异染色质中起着额外的控制作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Single-cell multi-omic detection of DNA methylation and histone modifications reconstructs the dynamics of epigenomic maintenance
DNA methylation and histone modifications encode epigenetic information. Recently, major progress was made to measure either mark at a single-cell resolution; however, a method for simultaneous detection is lacking, preventing study of their interactions. Here, to bridge this gap, we developed scEpi2-seq. Our technique provides a readout of histone modifications and DNA methylation at the single-cell and single-molecule level. Application in a cell line with the FUCCI cell cycle reporter system reveals how DNA methylation maintenance is influenced by the local chromatin context. In addition, profiling of H3K27me3 and DNA methylation in the mouse intestine yields insights into epigenetic interactions during cell type specification. Differentially methylated regions also demonstrated independent cell-type regulation in addition to H3K27me3 regulation, which reinforces that CpG methylation acts as an additional layer of control in facultative heterochromatin. This work presents scEpi2-seq, a method for simultaneous single-cell profiling of DNA methylation and histone modifications, enabling direct investigation of the interplay between these two epigenomic marks.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


