通过iswi强制核小体分期控制髓系谱系保真度和对刺激的反应
IF 26.3
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
摘要
染色质重塑子和先驱转录因子(tf)之间的相互作用调节顺式调控元件的可及性,以维持细胞身份和转录保真度。我们研究了模拟开关(ISWI)染色质重塑酶(核小体间距的关键调节因子)对巨噬细胞分化和激活的影响,重点研究了骨髓细胞中唯一的ISWI atp酶SMARCA5。骨髓源性巨噬细胞中条件性Smarca5的缺失破坏了PU.1结合位点附近的核小体分期,PU.1是骨髓特性所必需的先导TF,但不改变PU.1的占用。然而,SMARCA5缺失增加了C/EBPβ(弱先导TF)结合基序的可及性,使其能够结合到非造血谱系中活跃的调控区域,并导致谱系不适当的转录。这些变化还增加了刺激诱导的tf结合位点的可及性,导致巨噬细胞过度激活和刺激不适宜基因的错误表达。因此,smarca5依赖的核小体相位抑制C/EBPβ和刺激诱导的TF结合,确保巨噬细胞谱系规范和激活过程中的转录保真度,在其他免疫细胞类型中可能具有类似的作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
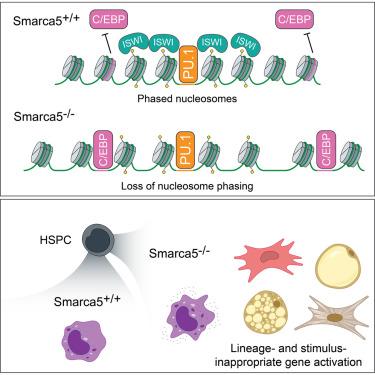
Control of myeloid lineage fidelity and response to stimuli by ISWI-enforced nucleosome phasing
The interplay between chromatin remodelers and pioneer transcription factors (TFs) regulates cis-regulatory element accessibility to maintain cell identity and transcriptional fidelity. We investigated the impact of imitation of switch (ISWI) chromatin remodelers, key regulators of nucleosome spacing, on macrophage differentiation and activation, focusing on SMARCA5, the sole ISWI ATPase in myeloid cells. Conditional Smarca5 deletion in bone marrow-derived macrophages disrupted nucleosome phasing near sites bound by PU.1, a pioneer TF essential for myeloid identity, without altering PU.1 occupancy. However, SMARCA5 loss increased accessibility at motifs bound by C/EBPβ, a weak pioneer TF, enabling binding to regulatory regions active in non-hematopoietic lineages and causing lineage-inappropriate transcription. These changes also increased accessibility at sites bound by stimulus-induced TFs, leading to macrophage hyperactivation and mis-expression of stimulus-inappropriate genes. Thus, SMARCA5-dependent nucleosome phasing restrains C/EBPβ and stimulus-induced TF binding, ensuring transcriptional fidelity during macrophage lineage specification and activation, with likely similar roles in other immune cell types.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Immunity
医学-免疫学
CiteScore
49.40
自引率
2.20%
发文量
205
审稿时长
6 months
期刊介绍:
Immunity is a publication that focuses on publishing significant advancements in research related to immunology. We encourage the submission of studies that offer groundbreaking immunological discoveries, whether at the molecular, cellular, or whole organism level. Topics of interest encompass a wide range, such as cancer, infectious diseases, neuroimmunology, autoimmune diseases, allergies, mucosal immunity, metabolic diseases, and homeostasis.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


