单细胞新生转录揭示了基因组的稀疏使用和可塑性
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
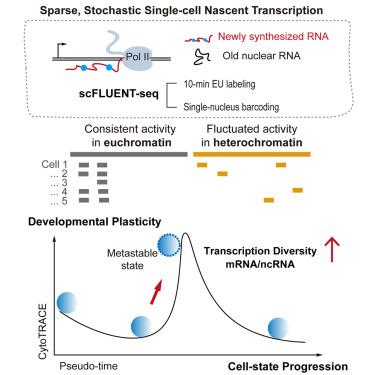
从一个共同的基因组理解细胞分化在后生动物需要单细胞转录分析。我们介绍了单细胞全长eu标记新生RNA测序(scFLUENT-seq),这是一种单细胞新生RNA测序方法,使用简短的10分钟代谢标记来捕获全基因组转录。令人惊讶的是,单个细胞-从脾淋巴细胞到多能干细胞-仅转录基因组的0.02%-3.1%,而不是80%,这揭示了有限的基因组参与和深刻的细胞类型和细胞间异质性。基因间转录,特别是来自异染色质的转录,是普遍和随机的。启动子相关的反义转录和基因转录很少同时发生在同一个细胞中。近端基因间转录包括基因读通和独立起始,而远端基因间转录在很大程度上独立于邻近基因,并与转录多样性增加相关,这是细胞可塑性的标志。尽管整体RNA合成和转换是大量耦合的,但单个mRNA转录和衰变在单个细胞中协调不佳,这表明存在噪声缓冲机制。总的来说,scFLUENT-seq揭示了单细胞异质性和状态转变背后的复杂编码和非编码转录动力学。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Single-cell nascent transcription reveals sparse genome usage and plasticity
Understanding cell diversification from a common genome in metazoans requires single-cell transcriptional analysis. We introduce single-cell full-length EU-labeled nascent RNA sequencing (scFLUENT-seq), a single-cell nascent RNA sequencing method using brief 10-min metabolic labeling to capture genome-wide transcription. Surprisingly, individual cells—from splenic lymphocytes to pluripotent stem cells—transcribe only ∼0.02%–3.1% of the genome, versus >80% in bulk, revealing limited genome engagement and profound cell-type and cell-to-cell heterogeneity. Intergenic transcription, especially from heterochromatin, is pervasive and stochastic. Promoter-associated antisense and genic transcription rarely co-occur in the same cell. Proximal intergenic transcription involves both gene readthrough and independent initiation, while distal intergenic transcription is largely independent of neighboring genes and correlates with increased transcriptional diversity, a hallmark of cellular plasticity. Although global RNA synthesis and turnover are coupled in bulk, individual mRNA transcription and decay are poorly coordinated in single cells, suggesting noise-buffering mechanisms. Overall, scFLUENT-seq uncovers complex coding and noncoding transcriptional dynamics that underlie single-cell heterogeneity and state transitions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


