空间蛋白质组学揭示了初级纤毛的内在异质性
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
初级纤毛是在大多数人体细胞中发现的重要细胞器。它们的功能障碍与具有广泛表型谱的遗传性纤毛病有关。尽管它们具有重要意义,但由于分析纤毛蛋白组成的局限性,纤毛在不同细胞类型中的具体作用仍然知之甚少。我们利用基于抗体的空间蛋白质组学将人类蛋白质图谱扩展到初级纤毛。我们的分析确定了三种细胞系中715个蛋白质的纤毛下位置,检查了128,156个纤毛。我们发现69%的纤毛蛋白质组具有细胞类型特异性,78%的纤毛具有单纤毛异质性。我们的研究结果将纤毛描述为传感器,调节它们的蛋白质组来有效地感知环境并计算细胞反应。我们揭示了91个纤毛蛋白,并在一个具有重叠纤毛病表型特征的临床病例中发现了CREB3的遗传候选变体。这个开放的、空间的纤毛图集促进了纤毛和纤毛病的研究。本文章由计算机程序翻译,如有差异,请以英文原文为准。
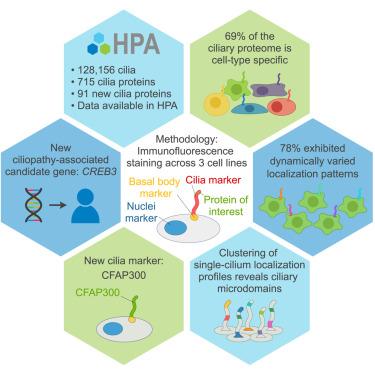
Intrinsic heterogeneity of primary cilia revealed through spatial proteomics
Primary cilia are critical organelles found on most human cells. Their dysfunction is linked to hereditary ciliopathies with a wide phenotypic spectrum. Despite their significance, the specific roles of cilia in different cell types remain poorly understood due to limitations in analyzing ciliary protein composition. We employed antibody-based spatial proteomics to expand the Human Protein Atlas to primary cilia. Our analysis identified the subciliary locations of 715 proteins across three cell lines, examining 128,156 individual cilia. We found that 69% of the ciliary proteome is cell-type specific, and 78% exhibited single-cilia heterogeneity. Our findings portray cilia as sensors tuning their proteome to effectively sense the environment and compute cellular responses. We reveal 91 cilia proteins and found a genetic candidate variant in CREB3 in one clinical case with features overlapping ciliopathy phenotypes. This open, spatial cilia atlas advances research on cilia and ciliopathies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


