基于微流控液滴角分辨光散射成像的多路、快速表型抗生素敏感性检测
IF 13
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
目前迫切需要一种能够评估微生物样品对多种抗生素敏感性的快速、高通量表型抗微生物药敏试验(AST)。本研究建立了一个多路快速AST平台,利用微流控技术进行高通量单细胞分析,利用二维角分辨光散射技术进行生长检测,并利用光纤荧光检测来识别每个微滴内的抗生素状况。方法采用荧光染料对多种抗生素条件进行编码,并用单细胞包膜,实现一次检测多种抗生素。我们利用卷积神经网络(cnn)和统计模型来评估各种金黄色葡萄球菌菌株的生长情况,并确定对不同抗生素的敏感性概率。结果该平台与圆盘扩散参考方法在孵育3小时后达到95%的分类一致性,表明与建立的VITEK 2系统对被测菌株和抗生素具有相同的准确性。值得注意的是,与VITEK 2相比,我们的平台将孵育时间缩短了5-11 h,与金标准圆盘扩散法相比缩短了13-17 h。结论我们的技术向实现抗生素耐药谱的真正表型测定迈出了一大步,为及时的抗菌药物治疗决策提供了依据。本文章由计算机程序翻译,如有差异,请以英文原文为准。
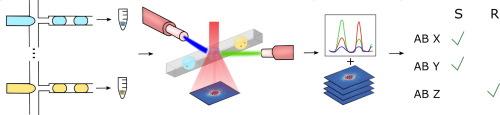
Multiplexed, rapid phenotypic antibiotic susceptibility testing based on angle-resolved light scattering imaging of microfluidic droplets
Introduction
There is an urgent need for rapid, high-throughput phenotypic antimicrobial susceptibility testing (AST) capable of assessing a microbial sample’s susceptibility to multiple antibiotics.Objectives
In this study, we have established a multiplexed rapid AST platform that employs droplet microfluidics for high-throughput single-cell based analysis, 2D angle-resolved light scattering for growth detection, and fluorescence detection via optical fibers to identify the antibiotic condition within each droplet.Methods
For this, multiple antibiotic conditions are coded with fluorescence dyes and encapsulated with single cells to enable the testing of multiple antibiotics in a single experiment. We utilize convolutional neural networks (CNNs) and statistical models to assess the growth of various Staphylococcus aureus strains and determine the probability of susceptibility to different antibiotics.Results
Our platform achieved a 95 % categorical agreement with the disc diffusion reference method after just three hours of incubation, demonstrating the same level of accuracy as the established VITEK 2 system for the tested strains and antibiotics. Notably, our platform reduced the incubation time by 5–11 h compared to VITEK 2 and by 13–17 h compared to the gold standard disc diffusion method.Conclusions
With the presented innovations, our technology takes a big step towards realizing true phenotypic determination of antibiotic resistance profiles for timely antimicrobial treatment decisions.求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


