多尺度蛋白质组学模型揭示了驱动阿尔茨海默病发病机制的蛋白质网络
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
阿尔茨海默病(AD)是最常见的痴呆形式,其发病机制的分子机制仍然知之甚少。蛋白质组学为阐明阿尔茨海默病的发病机制提供了一种至关重要的方法,因为蛋白质表达的改变比遗传或转录组学水平的变化更直接地与表型结果相关。在这项研究中,我们通过整合AD易感大脑区域的大规模匹配蛋白质组学和遗传数据,开发了AD的多尺度蛋白质组学网络模型。这些模型揭示了详细的蛋白质相互作用结构,并确定了参与AD进展的推定关键驱动蛋白(kdp)。值得注意的是,网络分析揭示了ad相关的子网络,该子网络捕获了神经胶质细胞之间的相互作用。AHNAK是胶质神经元网络中的顶级KDP,在基于人类诱导多能干细胞(iPSC)的AD模型中得到了实验验证。这种对失调蛋白调控网络和kdp的系统鉴定为开发AD的创新治疗策略奠定了基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
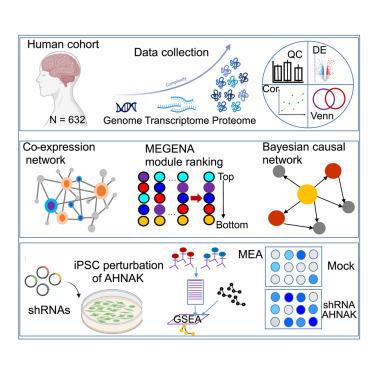
Multiscale proteomic modeling reveals protein networks driving Alzheimer’s disease pathogenesis
The molecular mechanisms underlying the pathogenesis of Alzheimer’s disease (AD), the most common form of dementia, remain poorly understood. Proteomics offers a crucial approach to elucidating AD pathogenesis, as alterations in protein expression are more directly linked to phenotypic outcomes than changes at the genetic or transcriptomic level. In this study, we develop multiscale proteomic network models for AD by integrating large-scale matched proteomic and genetic data from brain regions vulnerable to the disease. These models reveal detailed protein interaction structures and identify putative key driver proteins (KDPs) involved in AD progression. Notably, the network analysis uncovers an AD-associated subnetwork that captures glia-neuron interactions. AHNAK, a top KDP in this glia-neuron network, is experimentally validated in human induced pluripotent stem cell (iPSC)-based models of AD. This systematic identification of dysregulated protein regulatory networks and KDPs lays down a foundation for developing innovative therapeutic strategies for AD.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


