多模态融合模型结合SERS光谱和临床病理特征预测乳腺癌新辅助治疗反应
IF 6
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
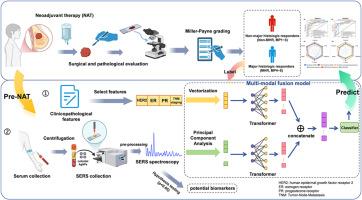
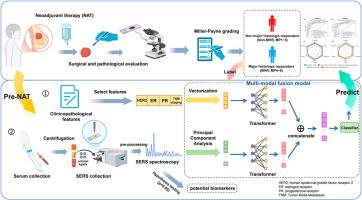
乳腺癌仍然是全球妇女健康的重大威胁,新辅助治疗(NAT)在治疗中起着关键作用。早期预测NAT疗效对于个性化治疗和改善患者预后至关重要。Miller-Payne (MP)分级系统是一种被广泛接受的评估治疗反应的标准,将患者分为非主要组织学反应者(MP1 ~ MP3)或主要组织学反应者(MP4 ~ MP5)。本研究建立了一种综合临床病理特征和治疗前血清表面增强拉曼光谱(SERS)数据的多模式融合模型,以预测乳腺癌患者的NAT反应。利用主成分分析(PCA)进行频谱降维,利用Transformer架构进行特征提取,该模型在训练队列上的准确率达到92.6%,明显优于仅使用SERS或临床病理特征的单模态模型。在独立队列上的双盲验证证实了该模型的泛化性,准确率为90%,受试者工作特征曲线下面积(AUC)为93%。SERS分析显示,尿酸、色氨酸、磷脂和胶原蛋白在光谱上存在显著差异,有可能作为NAT疗效预测的生物标志物。本研究创新性地将血清SERS数据与临床病理特征相结合,预测乳腺癌患者的NAT反应。多模态融合模型,通过PCA和Transformer架构增强,捕获生物分子和临床信息,提高预测精度和鲁棒性。这种无创、经济高效的工具使临床医生能够避免无效的NAT,优化治疗策略,改善患者的预后。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Multi-modal fusion model combines SERS spectroscopy and clinicopathological features to predict neoadjuvant therapy response in breast cancer
Breast cancer remains a significant global health threat to women, with neoadjuvant therapy (NAT) playing a critical role in treatment. Early prediction of NAT efficacy is essential for personalizing therapy and improving patient outcomes. The Miller-Payne (MP) grading system is a widely accepted standard for evaluating treatment response, categorizing patients as non-major histologic responders (MP1∼MP3) or major histologic responders (MP4∼MP5). This study developed a multi-modal fusion model integrating clinicopathological features and pre-treatment serum surface-enhanced Raman spectroscopy (SERS) data to predict NAT response in breast cancer patients. Leveraging Principal Component Analysis (PCA) for spectral dimensionality reduction and a Transformer architecture for feature extraction, the model achieved an accuracy of 92.6 % on the training cohort, significantly outperforming single-modal models using only SERS or clinicopathological features. Double-blind validation on an independent cohort confirmed the model's generalizability with an accuracy of 90 % and an area under the receiver operating characteristic curve (AUC) of 93 %. SERS analysis revealed significant spectral differences related to uric acid, tryptophan, phospholipids, and collagen, which have potential as biomarkers for NAT efficacy prediction. This study innovatively combined serum SERS data with clinicopathological features to predict NAT response in breast cancer patients. The multi-modal fusion model, enhanced by PCA and a Transformer architecture, captured biomolecular and clinical information, improving prediction accuracy and robustness. This non-invasive, cost-effective tool enables clinicians to avoid ineffective NAT, optimize treatment strategies, and improve patient outcomes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


