灵活性还是不确定性?AlphaFold 2 pLDDT的关键评估
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
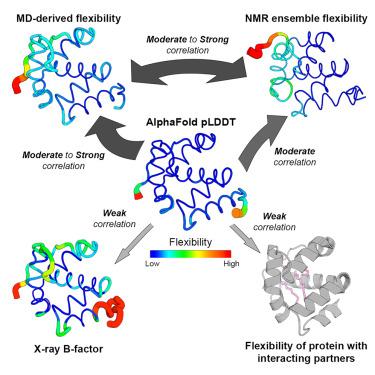
AlphaFold 2的发布和随后AlphaFold 3的开发对蛋白质结构预测产生了深远的影响,提供了接近实验的精度。然而,AF2的信心指数(pLDDT)作为蛋白质柔韧性指标的效用仍未得到充分的探索和争论。在这项大规模研究中,我们通过将AF2的pLDDT与来自ATLAS数据集、NMR集合和实验b因子的分子动力学(MD)模拟得出的灵活性指标进行比较,评估了AF2的pLDDT作为蛋白质灵活性的预测因子。在这种情况下,我们还评估了ESMFold pLDDT和AlphaFold 3的效率。我们的研究结果表明,AF2 pLDDT与MD和核磁共振衍生的灵活性指标有合理的相关性,但在相互作用的伙伴存在时无法捕获灵活性,因此需要谨慎解释。此外,与b因子值相比,AF2 pLDDT在评估蛋白质柔韧性方面似乎更相关。虽然AF3在捕获蛋白质动力学方面略有改进,但MD模拟在综合灵活性评估方面仍然优越。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Flexibility or uncertainty? A critical assessment of AlphaFold 2 pLDDT
Release of AlphaFold 2 and subsequent development of AlphaFold 3 had a profound impact on protein structure prediction, providing near-experimental accuracy. However, the utility of AF2’s confidence index (pLDDT) as indicators of protein flexibility remains underexplored and debated. In this large-scale study, we evaluate AF2’s pLDDT as a predictor of protein flexibility by comparing it with flexibility metrics derived from molecular dynamics (MD) simulations from the ATLAS dataset, NMR ensembles, and experimental B-factors. We also assess the efficiency of ESMFold pLDDT and AlphaFold 3 in this context. Our findings reveal that AF2 pLDDT reasonably correlates with MD and NMR-derived flexibility metrics, but fails to capture flexibility in the presence of interacting partners, and therefore need to be cautiously interpreted. Furthermore, AF2 pLDDT appears more relevant than B-factor values for evaluation of protein flexibility. While AF3 shows slight improvements in capturing protein dynamics, MD simulations remain superior for comprehensive flexibility assessment.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


