丁香假单胞菌avrPto5的功能研究。猕猴桃科及其与调节植物防御的锌指蛋白的相互作用
IF 3.3
3区 农林科学
Q2 PLANT SCIENCES
引用次数: 0
摘要
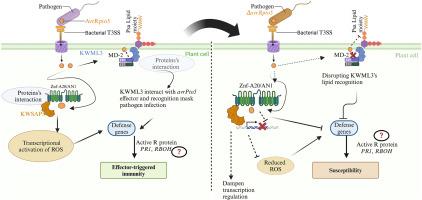
丁香假单胞菌。猕猴桃菌是影响猕猴桃的重要病原菌,不同菌株的毒力不同。这些致病菌利用III型效应蛋白(T3Es)作为必要的毒力因子来抑制宿主免疫并引起疾病。然而,在许多情况下,对T3Es的分子功能的研究仍然较少。在这项研究中,效应蛋白AvrPto5的缺失与丁香假单胞菌pv。比较猕猴桃野生型菌株G1,解码其致病机制。当渗透到猕猴桃品种“红羊”的叶片和树枝时,ΔavrPto5基因敲除突变株比野生型菌株G1表现出更大的病变长度和更大的疾病面积,表明突变增强了毒力。进一步通过酵母双杂交(Y2H)对猕猴桃基因组文库进行筛选,发现avrPto5与猕猴桃寄主KWSAP5和KWML3两个逆境相关蛋白(SAP)之间存在直接相互作用,并通过GST下拉实验验证了这一结果。KWSAP5是A20/AN1锌指蛋白家族成员,与拟南芥中的AT3G12630密切相关,含有保守的Znf-A20和zf-AN1结构域,参与氧化应激和抗丁香卟啉感染。Y2H和gst下拉实验强调了avrPto5在调节宿主-病原体相互作用中的作用,其缺失可能会破坏正常的免疫信号通路,导致致病性增加。本研究提高了对丁香假单胞菌的分子决定因素的认识。并强调了avrPto5与宿主SAP蛋白之间的关键相互作用,为开发抗猕猴桃品种提供了潜在的靶点。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Functional insights into avrPto5 from Pseudomonas syringae pv. actinidiae and its interaction with zinc finger proteins regulating plant defenses
Pseudomonas syringae pv. actinidiae is a significant pathogen affecting kiwifruit, with varying degrees of virulence among its strains. These pathogenic bacteria use type III effector proteins (T3Es) as essential virulence factors to suppress host immunity and cause disease. However, in many cases, the molecular function of T3Es remains less studied. In this study, a deletion of the effector protein AvrPto5 with the P. syringae pv. actinidiae wild-type strain G1 was compared to decode its pathogenic mechanisms. Upon infiltration into leaves and branches of the kiwifruit cultivar “Hongyang”, the ΔavrPto5 knockout mutant strain exhibited increased lesion lengths and larger disease areas compared to the wild-type strain G1, indicating enhanced virulence due to the mutation. Further investigation into kiwifruit genome mediated library screening through yeast two-hybrid (Y2H) indicated direct interactions between avrPto5 and two stress associated proteins (SAP) of kiwifruit host, KWSAP5 and KWML3, and validated by GST pull-down assays. This work was primarily focused on KWSAP5, a member of the A20/AN1 zinc finger protein family, closely related to AT3G12630 in Arabidopsis thaliana, containing conserved Znf-A20 and zf-AN1 domains and involved in oxidative stress and resistance against P. syringae infection. Y2H and GST-pull-down assays underscore avrPto5 role in modulating host-pathogen interactions, where its absence may disrupt normal immune signaling pathways, leading to increased pathogenicity. This study enhances the understanding of the molecular determinants of P. syringae pv. actinidiae virulence and emphasizes the critical interaction between avrPto5 and host SAP proteins, presenting potential targets for the development of resistant kiwifruit cultivars.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
4.30
自引率
7.40%
发文量
130
审稿时长
38 days
期刊介绍:
Physiological and Molecular Plant Pathology provides an International forum for original research papers, reviews, and commentaries on all aspects of the molecular biology, biochemistry, physiology, histology and cytology, genetics and evolution of plant-microbe interactions.
Papers on all kinds of infective pathogen, including viruses, prokaryotes, fungi, and nematodes, as well as mutualistic organisms such as Rhizobium and mycorrhyzal fungi, are acceptable as long as they have a bearing on the interaction between pathogen and plant.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


