桥接成像和基因组学:领域知识引导空间转录组学分析
IF 15.5
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
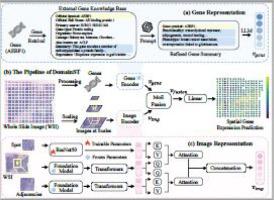
空间转录组学(ST)提供空间分辨率的基因表达分布映射到高分辨率的全幻灯片图像(WSIs),揭示细胞形态和基因表达谱之间的关联。然而,与ST数据收集相关的高成本和设备限制导致了ST数据集的稀缺。此外,现有的ST数据集往往表现出稀疏的基因表达分布,这限制了基于wsi的基因表达预测模型的准确性和泛化性。为了应对这些挑战,我们提出了DomainST(领域知识引导的空间转录组学分析),这是一个新的框架,通过大语言模型(LLMs)利用领域知识提取有效的基因表示,并利用基础模型获得鲁棒的图像特征,以增强空间基因表达预测。具体而言,我们利用公共基因参考数据库检索全面的基因摘要,并使用法学硕士来完善基因描述并生成信息丰富的基因嵌入。同时,我们应用医学视觉语言基础模型提取多尺度的鲁棒图像表示,捕捉wsi的空间背景。我们进一步设计了一个多模态混合专家融合模块,以有效地整合多模态数据,利用跨模态的互补信息。在三个公共ST数据集上进行的大量实验表明,我们的方法始终优于最先进的(SOTA)方法,与SOTA相比,在所有数据集上PCC@50的增长幅度从6.7%到13.7%不等,证明了将基础模型和llm衍生的领域知识结合起来进行基因表达预测的有效性。我们的代码和基因功能可以在https://github.com/coffeeNtv/DomainST上获得。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Bridging imaging and genomics: Domain knowledge guided spatial transcriptomics analysis
Spatial Transcriptomics (ST) provides spatially resolved gene expression distributions mapped onto high-resolution Whole Slide Images (WSIs), revealing the association between cellular morphology and gene expression profiles. However, the high costs and equipment constraints associated with ST data collection have led to a scarcity of ST datasets. Moreover, existing ST datasets often exhibit sparse gene expression distributions, which limit the accuracy and generalizability of gene expression prediction models derived from WSIs. To address these challenges, we propose DomainST (Domain knowledge-guided Spatial Transcriptomics analysis), a novel framework that leverages domain knowledge through Large Language Models (LLMs) to extract effective gene representations and utilizes foundation models to obtain robust image features for enhanced spatial gene expression prediction. Specifically, we utilize public gene reference databases to retrieve comprehensive gene summaries and employ LLMs to refine gene descriptions and generate informative gene embeddings. Concurrently, we apply medical visual-language foundation models to distill robust image representations at multiple scales, capturing the spatial context of WSIs. We further design a multimodal mixture of experts fusion module to effectively integrate multimodal data, leveraging complementary information across modalities. Extensive experiments conducted on three public ST datasets indicate that our method consistently outperforms state-of-the-art (SOTA) methods, with increases ranging from 6.7 % to 13.7 % in PCC@50 across all datasets compared to the SOTA, demonstrating the effectiveness of combining foundation models and LLM-derived domain knowledge for gene expression prediction. Our code and gene features are available at https://github.com/coffeeNtv/DomainST.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Information Fusion
工程技术-计算机:理论方法
CiteScore
33.20
自引率
4.30%
发文量
161
审稿时长
7.9 months
期刊介绍:
Information Fusion serves as a central platform for showcasing advancements in multi-sensor, multi-source, multi-process information fusion, fostering collaboration among diverse disciplines driving its progress. It is the leading outlet for sharing research and development in this field, focusing on architectures, algorithms, and applications. Papers dealing with fundamental theoretical analyses as well as those demonstrating their application to real-world problems will be welcome.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


