长链非编码rna的全基因组鉴定和功能预测
IF 4.1
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
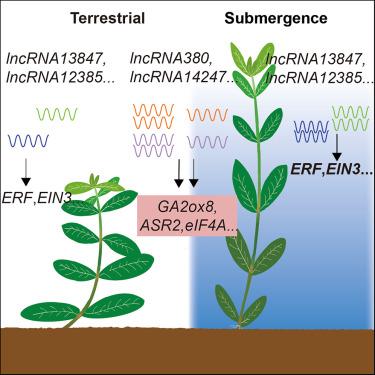
植物有一种非凡的能力来改变它们的生长和发育,以适应波动的环境。表观遗传学在调节塑性表型中起着关键作用。基于长读PacBio测序和时间过程基因表达谱,研究人员对水浸诱导塑料生长的lncrna进行了全基因组调查和功能预测。共鉴定出141个水下响应lncrna,并对其顺式和反式靶向基因进行了预测。lncRNA靶点的功能注释和验证表明,lncRNA14247、lncRNA13847和lncRNA12385通过介导淹没时各种植物激素间的串扰,在植物激素稳态中发挥重要作用,从而影响可塑性发育。结果还揭示了lncRNA13847和lncRNA7566通过靶向光信号成分(如pif)介导淹没诱导的生长改变的潜在作用。这些发现突出了lncrna在淹没反应中的重要性,并为黄叶草表型可塑性的分子基础提供了见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Genome-wide identification and functional prediction of long non-coding RNAs in plastic growth in Alternanthera philoxeroides
Plants have a remarkable ability to alter their growth and development to adapt to fluctuating environments. Epigenetics plays a pivotal role in regulating plastic phenotypes. Based on long-read PacBio sequencing and time-course gene expression profiling, a genome-wide survey and functional prediction of lncRNAs potentially involved in submergence-induced plastic growth was conducted in A. philoxeroides. A total of 141 submergence-responsive lncRNAs were identified, with their cis- and trans-targeting genes being predicted. Functional annotation and validation of lncRNA targets indicated the significant roles of lncRNA14247, lncRNA13847, and lncRNA12385 on phytohormone homeostasis by mediating crosstalk between various phytohormones upon submergence, and thereby impacting plastic development. The results also unveiled the potential roles of lncRNA13847 and lncRNA7566 in mediating submergence-induced growth alterations by targeting light signal components, such as PIFs. These findings highlighted the significance of lncRNAs in submergence responses and provided insights into the molecular underpinnings of phenotypic plasticity in A. philoxeroides.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


