单细胞多组学揭示了胚胎脂肪尾形态发生过程中的多种脂肪形成途径和多种多谱系特化
IF 4.1
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
由于时间敏感性、有限的组织体积和伦理问题,胚胎脂肪形成仍然是哺乳动物脂肪生物学中最不为人所知的方面之一。在这里,我们独特地将单细胞多组学测序应用于胖尾羊的发育脂肪组织,其特征是遗传决定的,在胚胎发生期间尾巴有显著的脂肪沉积。我们的数据集涵盖了脂肪形成的所有阶段(E50至E80),揭示了脂肪沉积的三个主要细胞来源:祖细胞和干细胞、结缔组织祖细胞和血管平滑肌细胞。通过整合scRNA-seq、snATAC-seq和功能验证,我们确定了控制脂肪形成的关键增强子驱动基因调控网络(egrn), DBI通过与PPARG的相互作用成为关键的调控因子。此外,我们描述了与脂肪形成相关的血管生成、骨生成、软骨生成和肌肉生成的发育轨迹和独特的egrn。我们的发现为哺乳动物胚胎脂肪形成提供了新的见解,并揭示了支配谱系特化的关键规律。本文章由计算机程序翻译,如有差异,请以英文原文为准。
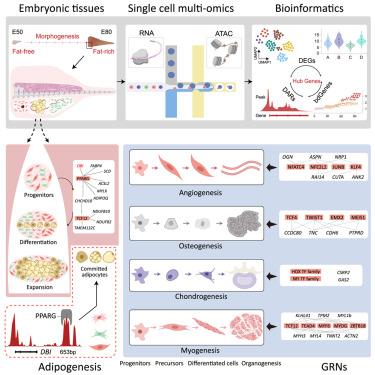
Single-cell multiomics reveals multiple adipogenic pathways and diverse multilineage specializations during embryonic fat tail morphogenesis
Embryonic adipogenesis remains one of the least understood aspects of adipose biology in mammals due to time sensitivity, limited tissue volume, and ethical concerns. Here, we uniquely applied single-cell multi-omics sequencing to the developing adipose tissues of fat-tailed sheep, characterized by genetically determined, significant fat deposition in the tail during embryogenesis. Our dataset spans all stages of adipogenesis (E50 to E80), revealing three major cellular origins of fat deposition: progenitor and stem cells, connective tissue progenitors, and vascular smooth muscle cells. By integrating scRNA-seq, snATAC-seq, and functional validation, we identified key enhancer-driven gene regulatory networks (eGRNs) governing adipogenesis, with DBI emerging as a critical regulator through its interaction with PPARG. Additionally, we delineated developmental trajectories and unique eGRNs underlying angiogenesis, osteogenesis, chondrogenesis, and myogenesis associated with fat formation. Our findings provide novel insights into embryonic adipogenesis in mammals and reveal critical regulons governing lineage specialization.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


