54种癌细胞系蛋白质组学特征的综合多维比较
IF 4.1
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
人类癌细胞系是生物医学研究中必不可少的模型系统。我们使用两种主要的蛋白质组学技术:质谱(MS)和反相蛋白质阵列(RPPA),对来自不同组织的54种广泛使用的癌细胞系进行了多层次蛋白质组学分析。我们的分析确定了10,088个蛋白,7,469个磷酸化蛋白中的33,161个磷酸化位点,5,966个糖蛋白中的14,228个糖基化位点上的56,320个位点特异性聚糖,以及305个药物相关蛋白和磷酸化蛋白靶点。对这一丰富数据集的分析产生了许多生物学见解,包括区分组织起源和细胞系特异性激酶激活模式的蛋白质特征,反映了癌症类型的信号多样性。这些发现可能为治疗策略提供信息,并支持合理的模型系统选择。此外,MS和RPPA显示出一致的折叠变化估计,并提供了蛋白质组和信号变异的互补观点。这个全面的资源促进了生物标志物的发现,信号分析和转化肿瘤学研究跨越不同的人类肿瘤类型。本文章由计算机程序翻译,如有差异,请以英文原文为准。
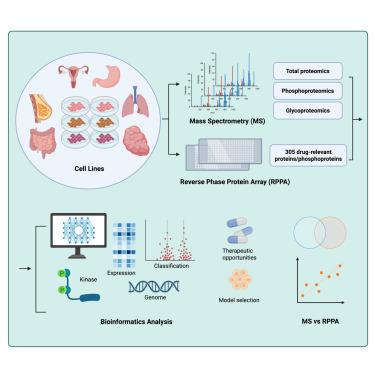
Integrated multi-dimensional comparison of proteomic profiles across 54 cancer cell lines
Human cancer cell lines are essential model systems in biomedical research. We conducted multi-level proteomics analyses on 54 widely used cancer cell lines derived from various tissues using two prominent proteomics technologies: mass spectrometry (MS) and reverse-phase protein array (RPPA). Our analysis identified 10,088 proteins, 33,161 phosphorylation sites across 7,469 phosphoproteins, and 56,320 site-specific glycans on 14,228 glycosylation sites from 5,966 glycoproteins, along with 305 drug-relevant protein and phosphoprotein targets. Analysis of this rich dataset yielded numerous biological insights, including protein features that distinguish tissue origins and cell line-specific kinase activation patterns, reflecting signaling diversity across cancer types. These findings may inform therapeutic strategies and support rational model system selection. Additionally, MS and RPPA showed consistent fold-change estimation and provided complementary views of proteome and signaling variation. This comprehensive resource facilitates biomarker discovery, signaling analysis, and translational oncology research across diverse human tumor types.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


