参考基因组的选择损害了群体遗传分析
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
描述自然种群的遗传变异对进化生物学至关重要;然而,许多非模式物种缺乏基因组资源。本文通过将灰狐(Urocyon cinereoargenteus)的全基因组序列数据映射到同种参考和两个异种犬科动物(狗和北极狐)的基因组,证明参考偏差显著影响种群基因组分析。对同种基因组的定位使读对提高了约5%,并检测到26%-32%的snp和33%-35%的单基因。核苷酸多样性估计值提高了30%以上,FST从0.189提高到0.197,有效种群大小估计值比同种参考高出30% - 60%。异种参考染色体末端的重组率变化可达3倍。重要的是,FST异常值检测明显不同,异源基因组鉴定出的独特异常窗口数是其两倍。这些发现突出了参考基因组选择的影响和同源基因组资源对准确进化推断的重要性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
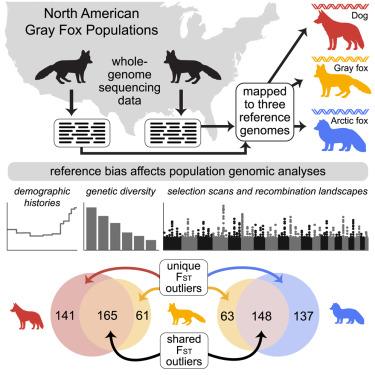
Reference genome choice compromises population genetic analyses
Characterizing genetic variation in natural populations is vital to evolutionary biology; however, many non-model species lack genomic resources. Here, we demonstrate that reference bias significantly affects population genomic analyses by mapping whole-genome sequence data from gray foxes (Urocyon cinereoargenteus) to a conspecific reference and two heterospecific canid genomes (dog and Arctic fox). Mapping to the conspecific genome improved read pairing by ∼5% and detected 26%–32% more SNPs and 33%–35% more singletons. Nucleotide diversity estimates increased by over 30%, FST increased from 0.189 to 0.197, and effective population size estimates were 30%–60% higher with the conspecific reference. Recombination rates varied by up to 3-fold at chromosome ends with heterospecific references. Importantly, FST outlier detection differed markedly, with heterospecific genomes identifying twice as many unique outlier windows. These findings highlight the impact of reference genome choice and the importance of conspecific genomic resources for accurate evolutionary inference.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


