YTHDF2通过m6a依赖性MTUS1/ATIP1 mRNA降解和线粒体失调驱动口腔鳞状细胞癌的进展。
IF 3.7
2区 生物学
Q2 CELL BIOLOGY
引用次数: 0
摘要
n6 -甲基腺苷(m6A)是最丰富的RNA修饰,通过YTHDF2等读取器蛋白调节mRNA的稳定性。在这里,我们研究YTHDF2在口腔鳞状细胞癌(OSCC)进展中的作用。临床分析显示,与正常组织相比,YTHDF2在OSCC肿瘤中的表达升高,与疾病晚期、转移和患者生存率降低有关。在机制上,OSCC表现出全局m6A低甲基化,而YTHDF2通过识别其3'非翻译区域的m6A基序,选择性地破坏肿瘤抑制MTUS1/ATIP1 mRNA的稳定性。使用慢病毒过表达和敲低模型的功能研究表明,YTHDF2促进肿瘤生长和线粒体功能障碍,而其抑制抑制恶性行为并稳定MTUS1/ATIP1。在体外和皮下异种移植模型中,共沉默MTUS1/ATIP1逆转了YTHDF2缺失的抗肿瘤作用。这些发现表明,YTHDF2通过m6a依赖性抑制MTUS1/ATIP1,作为OSCC进展的驱动因素,将线粒体失调与肿瘤发生联系起来。我们的研究提出了靶向治疗YTHDF2-MTUS1/ATIP1轴以改善OSCC的管理。本文章由计算机程序翻译,如有差异,请以英文原文为准。
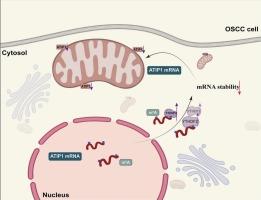
YTHDF2 drives oral squamous cell carcinoma progression via m6A-dependent degradation of MTUS1/ATIP1 mRNA and mitochondrial dysregulation
N6-methyladenosine (m6A), the most abundant RNA modification, regulates mRNA stability through reader proteins such as YTHDF2. Here, we investigate YTHDF2's role in oral squamous cell carcinoma (OSCC) progression. Clinical analyses revealed elevated YTHDF2 expression in OSCC tumors compared to normal tissues, correlating with advanced disease stage, metastasis, and reduced patient survival. Mechanistically, OSCC exhibits global m6A hypomethylation, while YTHDF2 selectively destabilizes tumor-suppressive MTUS1/ATIP1 mRNA by recognizing m6A motifs in its 3′ untranslated region. Functional studies using lentiviral overexpression and knockdown models demonstrated that YTHDF2 promotes tumor growth and mitochondrial dysfunction, whereas its suppression inhibits malignant behaviors and stabilizes MTUS1/ATIP1. Co-silencing MTUS1/ATIP1 reversed the anti-tumor effects of YTHDF2 depletion in vitro and in subcutaneous xenograft models. These findings establish YTHDF2 as a driver of OSCC progression through m6A-dependent suppression of MTUS1/ATIP1, linking mitochondrial dysregulation to tumorigenesis. Our study proposes therapeutic targeting of the YTHDF2-MTUS1/ATIP1 axis to improve OSCC management.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Cellular signalling
生物-细胞生物学
CiteScore
8.40
自引率
0.00%
发文量
250
审稿时长
27 days
期刊介绍:
Cellular Signalling publishes original research describing fundamental and clinical findings on the mechanisms, actions and structural components of cellular signalling systems in vitro and in vivo.
Cellular Signalling aims at full length research papers defining signalling systems ranging from microorganisms to cells, tissues and higher organisms.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


