小鼠胚胎发育过程中h3k9me2修饰的染色质区域的重组。
IF 2.1
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
摘要
细胞核中染色质的三维结构对调控基因表达至关重要。有两类由抑制染色质修饰组蛋白H3赖氨酸9二甲基(H3K9me2)划定并富集于核周围的大基因组区域:层相关结构域(LADs);和H3K9me2-only结构域(KODs),这些结构域与层接触最少,转录增强子高度富集。已经在多种细胞类型中研究了LADs。相比之下,KODs仅在多能性细胞中有特征,它们是固定的还是根据细胞类型重新排列仍有待确定。对不同胚胎小鼠组织的kod的分析表明,kod采用与谱系特异性增强子活性变化相关的细胞类型特异性配置。在KODs中,H3K9me2的局部细胞类型特异性缺失在活性谱系特异性增强子的H3K27ac峰上富集。KODs还在超长调控区域中富集,这表明KODs在基因的远程调控中起着重要作用。这些结果表明,kod是细胞类型特异性的,并维持细胞类型特异性增强子处于抑制状态,以允许组织和阶段特异性基因激活。本文章由计算机程序翻译,如有差异,请以英文原文为准。
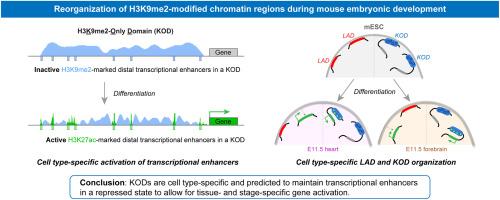
Reorganization of H3K9me2-modified chromatin regions during mouse embryonic development
The three-dimensional organization of chromatin in the nucleus is critical in regulating gene expression. There are two classes of large genomic regions demarcated by the repressive chromatin modification histone H3 lysine 9 dimethyl (H3K9me2) and enriched at the nuclear periphery: lamina-associated domains (LADs); and H3K9me2-only domains (KODs) which have minimal lamina contact and are highly enriched for transcriptional enhancers. LADs have been studied in multiple cell types. In contrast, KODs have been characterized only in pluripotent cells and it remains to be determined whether they are fixed or rearrange according to cell type. Analysis of KODs from various embryonic mouse tissues revealed that KODs adopt cell type-specific configurations that correlate with changes in lineage-specific enhancer activity. Within KODs, local cell type-specific depletion of H3K9me2 was enriched for H3K27ac peaks at active lineage-specific enhancers. KODs were also enriched across ultra-long regulatory regions suggesting a role for KODs in long-range gene regulation. These results suggest that KODs are cell-type specific and maintain cell type-specific enhancers in a repressed state to allow for tissue- and stage-specific gene activation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


