基因敲除提示小鼠后肢发育中存在Isl1增强子冗余。
IF 2.1
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
摘要
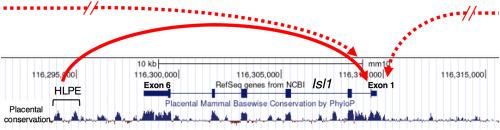
Isl1编码一个LIM同源结构域转录因子,该因子在侧板中胚层的后肢祖细胞中表达,是启动后肢发育所必需的。先前在小鼠胚胎中转基因lacZ的研究发现了位于Isl1基因3'的顺式元件,该元件可驱动lacZ报告基因在后肢祖细胞和鳃弓外胚层的表达。我们将这种顺式元件称为Isl1后肢祖增强子(HLPE)。我们前期的研究也发现转录因子SALL4在Isl1的HLPE富集,提示SALL4可能通过Isl1的HLPE调控Isl1的表达。我们试图确定Isl1 HLPE是否调节Isl1的表达,并通过CRISPR/Cas9创建缺乏Isl1 HLPE序列的突变小鼠。与野生型小鼠杂交后建立的Isl1 HLPE-/-小鼠系,除了长骨比野生型短外,没有出现明显的形态学缺陷。在妊娠中期观察到较短的长骨表型,并与几种软骨形成基因的表达失调有关,这表明HLPE的缺失会影响软骨形成。尽管怀疑Sall4通过Isl1 HLPE调控Isl1,但骨骼分析并未显示Isl1 HLPE-/-和条件Sall4突变之间有任何协同作用。原位杂交显示Isl1及其下游基因Tbx4在Isl1 HLPE-/-、tcr中表达正常;Sall4fl/fl突变体和Isl1 HLPE-/-;TCre;Sall4fl / fl突变体。最后,通过定量基因表达分析,Isl1在HLPE-/-和Isl1+/-中表达减少但未完全消除;Isl1 HLPE+/-胚,与野生型胚比较。同样,免疫染色后的定量成像分析显示,ISL1 HLPE-/-胚胎的鳃弓外胚层中ISL1信号减少。这些结果支持了我们的观点,即Isl1 HLPE序列在后肢祖细胞和鳃弓外胚层细胞中作为Isl1表达的增强子,并表明多个冗余增强子共同调节Isl1的表达。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Genetic knockout suggests Isl1 enhancer redundancy in mouse hindlimb development
Isl1 encodes a LIM homeodomain transcription factor, which is expressed in hindlimb progenitor cells in the lateral plate mesoderm and is required for initiating hindlimb development. Previous studies by lacZ transgenesis in mouse embryos identified a cis-element located 3’ to the Isl1 gene, which could drive lacZ reporter expression in hindlimb progenitor cells and the branchial arch ectoderm. We refer to this cis-element as the Isl1 hindlimb progenitor enhancer (HLPE). Our previous study also showed that SALL4, a transcription factor, is enriched at the Isl1 HLPE, suggesting that SALL4 may regulate Isl1 expression through Isl1 HLPE. We sought to determine whether Isl1 HLPE regulates Isl1 expression and created mutant mice that lack the Isl1 HLPE sequence by CRISPR/Cas9. The Isl1 HLPE−/− mouse lines, established after breeding with wild-type mice, did not exhibit gross morphological defects, except that their long bones are shorter than those of wild type. The shorter long bone phenotype was observed in the mid-gestation stage and was associated with misregulation of the expression of several chondrogenic genes, suggesting that the deletion of HLPE affects chondrogenesis. Although Sall4 regulation of Isl1 through Isl1 HLPE was suspected, skeletal analysis did not exhibit any synergy between Isl1 HLPE−/− and conditional Sall4 mutation. In situ hybridization showed seemingly normal expression of Isl1 and its downstream gene Tbx4 in Isl1 HLPE−/−, TCre; Sall4fl/fl mutants, and Isl1 HLPE−/−; TCre; Sall4fl/fl mutants. Finally, by quantitative gene expression analysis, Isl1 expression is reduced but not abolished in Isl1 HLPE−/− and Isl1+/−; Isl1 HLPE ± embryos, compared to wild-type embryos. Similarly, quantitative imaging analysis after immunostaining showed reduced ISL1 signals in the branchial arch ectoderm in Isl1 HLPE−/− embryos. These results support our notion that the Isl1 HLPE sequence functions as an enhancer for Isl1 expression in hindlimb progenitor cells and branchial arch ectoderm cells and suggest that multiple redundant enhancers co-regulate Isl1 expression.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


