利用蛋白质序列数据预测微生物代谢中的碳氮源偏好。
IF 1.9
4区 生物学
Q4 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
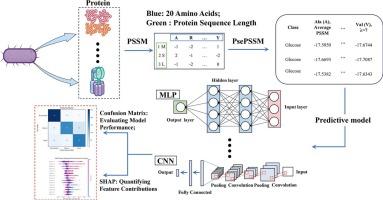
微生物精细化培养技术在环境污染物修复领域具有重要的应用价值。微生物碳氮需求的精确量化是优化培养条件和提高微生物生长和生产力的关键。本研究旨在利用深度学习算法探索微生物蛋白质序列与其特定营养需求(如碳源和氮源类型以及最佳碳氮比)之间的内在关系。从包括Ensembl Bacteria、DSMZ和NCBI Protein在内的权威数据库中收集了432种微生物和61种培养基配方。在数据分析中,利用位置特异性评分矩阵(PSSM)和伪位置特异性评分矩阵(PsePSSM)将微生物蛋白序列转换为高维数值特征矩阵,然后进行降维。采用多种机器学习算法构建微生物C/N源利用率预测模型。在分类任务中,C/N预测效果最好,准确率为99.60 %,其次是碳源预测,准确率为82.76 %,氮源预测准确率为70.05 %,说明微生物C/N需要量与其相关蛋白序列之间存在较强的相关性。此外,使用SHapley加性解释(SHAP)框架来分析特征贡献,增强了模型的可解释性。本研究的主要贡献在于提出了一种结合蛋白质功能注释和基于序列的特征提取预测微生物营养需求的集成框架,从而为优化微生物培养条件提供了新的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Prediction of carbon and nitrogen source preferences in microbial metabolism using protein sequence data
Microbial precision cultivation technology holds significant application value in the field of environmental pollutant remediation. Precise quantification of microbial carbon and nitrogen requirements is critical for optimizing culture conditions and enhancing microbial growth and productivity. This study aims to explore the intrinsic relationship between microbial protein sequences and their specific nutritional requirements (e.g., the types of carbon and nitrogen sources as well as the optimal carbon-to‑nitrogen (C/N) ratio) using deep learning algorithms. A total of 432 microbial species and 61 culture media formulations were collected from authoritative databases, including Ensembl Bacteria, DSMZ, and NCBI Protein. For data analysis, microbial protein sequences were converted into high-dimensional numerical feature matrices using the Position-Specific Scoring Matrix (PSSM) and Pseudo Position-Specific Scoring Matrix (PsePSSM), followed by dimensionality reduction. Multiple machine learning algorithms were employed to construct predictive models for microbial C/N source utilization. Among the classification tasks, C/N ratio prediction performed best, with an accuracy of 99.60 %, followed by carbon source prediction with an accuracy of 82.76 % and nitrogen source prediction with an accuracy of 70.05 %, suggesting a strong correlation between microbial C/N requirements and their associated protein sequences. Furthermore, model interpretability was enhanced using the SHapley Additive exPlanations (SHAP) framework to analyze feature contributions. The primary contribution of this study lies in proposing an integrated framework that combines protein function annotation with sequence-based feature extraction for predicting microbial nutritional requirements, thereby offering new insights for optimizing microbial culture conditions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of microbiological methods
生物-生化研究方法
CiteScore
4.30
自引率
4.50%
发文量
151
审稿时长
29 days
期刊介绍:
The Journal of Microbiological Methods publishes scholarly and original articles, notes and review articles. These articles must include novel and/or state-of-the-art methods, or significant improvements to existing methods. Novel and innovative applications of current methods that are validated and useful will also be published. JMM strives for scholarship, innovation and excellence. This demands scientific rigour, the best available methods and technologies, correctly replicated experiments/tests, the inclusion of proper controls, calibrations, and the correct statistical analysis. The presentation of the data must support the interpretation of the method/approach.
All aspects of microbiology are covered, except virology. These include agricultural microbiology, applied and environmental microbiology, bioassays, bioinformatics, biotechnology, biochemical microbiology, clinical microbiology, diagnostics, food monitoring and quality control microbiology, microbial genetics and genomics, geomicrobiology, microbiome methods regardless of habitat, high through-put sequencing methods and analysis, microbial pathogenesis and host responses, metabolomics, metagenomics, metaproteomics, microbial ecology and diversity, microbial physiology, microbial ultra-structure, microscopic and imaging methods, molecular microbiology, mycology, novel mathematical microbiology and modelling, parasitology, plant-microbe interactions, protein markers/profiles, proteomics, pyrosequencing, public health microbiology, radioisotopes applied to microbiology, robotics applied to microbiological methods,rumen microbiology, microbiological methods for space missions and extreme environments, sampling methods and samplers, soil and sediment microbiology, transcriptomics, veterinary microbiology, sero-diagnostics and typing/identification.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


