gpu加速同源搜索与MMseqs2。
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
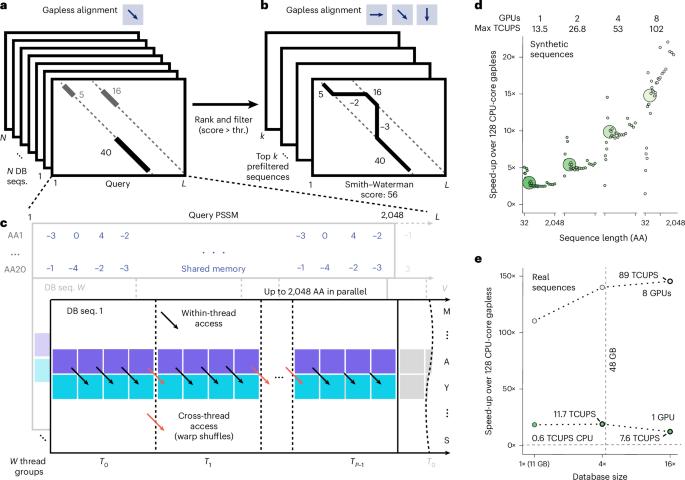
快速增长的蛋白质数据库需要更灵敏的搜索工具。在这里,图形处理单元(GPU)加速的MMseqs2提供了比2 × 64核CPU方法快6倍的单蛋白搜索速度,以前需要大量蛋白质批量。对于较大的查询批处理,它是最经济有效的解决方案,使用8个gpu,性能比最快的替代方法高出2.4倍。与标准AlphaFold2管道相比,ColabFold的蛋白质结构预测速度提高了31.8倍,与Foldseek的蛋白质结构搜索速度提高了4- 27x。MMseqs2-GPU在开源许可下可在https://mmseqs.com/获得。本文章由计算机程序翻译,如有差异,请以英文原文为准。
GPU-accelerated homology search with MMseqs2
Rapidly growing protein databases demand faster sensitive search tools. Here the graphics processing unit (GPU)-accelerated MMseqs2 delivers 6× faster single-protein searches than CPU methods on 2 × 64 cores, speeds previously requiring large protein batches. For larger query batches, it is the most cost-effective solution, outperforming the fastest alternative method by 2.4-fold with eight GPUs. It accelerates protein structure prediction with ColabFold 31.8× over the standard AlphaFold2 pipeline and protein structure search with Foldseek by 4–27×. MMseqs2-GPU is available under an open-source license at https://mmseqs.com/ . Graphics processing unit-accelerated MMseqs2 offers tremendous speedups for homology retrieval from metagenomic databases, query-centered multiple sequence alignment generation for structure prediction, and structural searches with Foldseek.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


