免疫应答过程中释放的长链不饱和脂肪酸刺激宿主-微生物跨界交流
IF 18.7
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
摘要
免疫反应可以显著改变肠道微生物群的结构和功能,导致共生微生物的快速转录和代谢变化。然而,参与这一过程的宿主介质及其对细菌的影响尚不清楚。在这里,使用鞭毛蛋白注射模型诱导免疫激活,我们发现不饱和长链脂肪酸(uLCFAs)作为广泛的调节剂被释放到肠腔并改变细菌基因表达。uLCFAs的腔内释放部分由宿主磷脂酶介导,包括PLA2G5。作为对uLCFAs的反应,蓝藻等共生体触发ohyA的表达,编码油酸水合酶,将有毒的uLCFAs转化为具有免疫调节特性的无毒羟基脂肪酸。值得注意的是,口服uLCFAs小鼠复制了许多由鞭毛蛋白诱导的细菌转录变化。这种分子环强调了宿主和微生物群之间复杂的相互作用,并揭示了免疫反应如何影响肠道共生功能。本文章由计算机程序翻译,如有差异,请以英文原文为准。
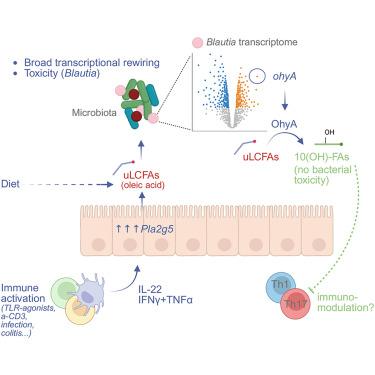
Long-chain unsaturated fatty acids released during immune responses stimulate host-microbe trans-kingdom communication
Immune responses can significantly alter the structure and function of the gut microbiota, leading to rapid transcriptional and metabolic shifts in commensal microbes. However, the host mediators involved in this process and their effects on bacteria remain poorly elucidated. Here, using a flagellin injection model to induce immune activation, we identified unsaturated long-chain fatty acids (uLCFAs) as broad modulators that are released into the gut lumen and alter bacterial gene expression. Luminal release of uLCFAs is partially mediated by host phospholipases, including PLA2G5. In response to uLCFAs, commensals such as Blautia trigger the expression of ohyA, encoding oleate hydratase, which converts toxic uLCFAs to non-toxic hydroxy fatty acids with immunomodulatory properties. Remarkably, oral administration of uLCFAs to mice replicates many of the bacterial transcriptional changes induced by flagellin. This molecular loop underscores the sophisticated interactions between host and microbiota and sheds light on how immune responses affect gut commensal functions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


