用于结核单核苷酸突变亚飞摩尔检测的dnazyme激活滚动圈扩增级联纳米机
IF 6
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
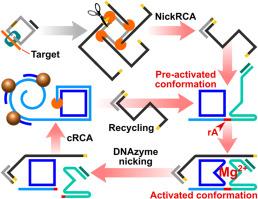
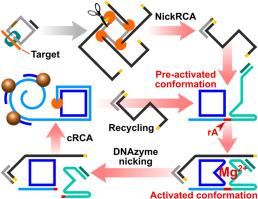
结核病(TB)仍然是一个全球性的健康危机,由于当前方法的高成本和性能限制,耐药菌株构成了重大的诊断挑战。耐利福平结核(RR-TB),其中约97%与rpoB 531T单核苷酸突变相关,是结核控制的关键靶点。基于挂锁探针连接的滚动圈扩增(RCA)能够区分单核苷酸变异,使其成为检测结核病耐药突变的一种有希望的方法。然而,基于挂锁探针连接的RCA只能达到亚皮摩尔的检测极限,限制了其应用。结果建立了一种用于结核分枝杆菌rpoB 531 (TCG - TTG)突变快速检测的多功能DNA纳米机。该纳米机器将dnazyme介导的裂解反应和RCA集成在一个空间受限的结构中。具体来说,通过将DNAzyme底物链、DNAzyme催化链、预形成的圆形模板和RCA引物共同组装成一个纳米结构,我们建立了一个有限的反应微环境,最大限度地减少了扩散距离,加速了反应动力学,同时有效地减少了级联步骤之间的串扰。利用DNA纳米机器的可编程性和检测探针功能化的磁性纳米颗粒进行实时光磁传感,我们的生物传感器实现了0.3 fM的检测限,总分析时间为100分钟,并展示了跨越6个数量级的动态检测范围。生物传感器的性能通过合成野生型序列、加标血清样本和临床痰DNA提取物进行验证。通过将级联DNA反应在空间上限制在单个纳米结构内,我们克服了复杂基质中均相级联反应的固有局限性,实现了自催化信号循环、RCA、纳米颗粒聚类和实时定量。这种方法为即时诊断RR-TB提供了一种可靠且具有成本效益的解决方案,并展示了检测其他单核苷酸突变的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
DNAzyme-activated rolling circle amplification cascade nanomachine for sub-femtomolar detection of tuberculosis single-nucleotide mutation
Background
Tuberculosis (TB) remains a global health crisis, with drug-resistant strains posing significant diagnostic challenges due to the high cost and performance limitations of current methods. Rifampicin-resistant TB (RR-TB), approximately 97% of which are associated with rpoB 531T single-nucleotide mutation, presents a critical target for TB control. Padlock probe ligation-based rolling circle amplification (RCA) enables discrimination of single-nucleotide variants, rendering it a promising approach for detecting TB drug-resistance mutations. However, padlock probe ligation-based RCA achieves only sub-picomolar limits of detection, limiting its application.
Results
Herein, we present a multifunctional DNA nanomachine designed for the rapid detection of the Mycobacterium tuberculosis (MTB) rpoB 531 (TCG to TTG) mutation. This nanomachine integrates a DNAzyme-mediated cleavage reaction and RCA within a spatially confined architecture. Specifically, by co-assembling a DNAzyme substrate strand, a DNAzyme catalytic strand, a preformed circular template, and an RCA primer into a single nanostructure, we established a confined reaction microenvironment that minimizes diffusion distances, enabling accelerated reaction kinetics while effectively reducing cross-talk between cascade steps. Leveraging the programmability of DNA nanomachines with detection probe-functionalized magnetic nanoparticles for real-time optomagnetic sensing, our biosensor achieved a detection limit of 0.3 fM with a total assay time of 100 min and exhibited a dynamic detection range spanning 6 orders of magnitude. The biosensor performance was validated using synthetic wild-type sequences, spiked serum samples, and clinical sputum DNA extracts.
Significance and novelty
By spatially confining cascade DNA reactions within a single nanostructure, we overcame intrinsic limitations of homogeneous cascade reactions in complex matrices, enabling autocatalytic signal recycling, RCA, nanoparticle clustering, and real-time quantification. This approach offers a robust and cost-effective solution for point-of-care RR-TB diagnosis and demonstrates the potential for detecting other single-nucleotide mutations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


