卡皮藻HMG-box-C1模块DNA识别的分子基础
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
HMG-box蛋白capicua (CIC)是一种保守的转录抑制因子,在发育和疾病中起关键作用。DNA的CIC结合既需要它的HMG-box,也需要一个叫做C1的单独结构域。这些结构域如何协同识别特定的DNA序列尚不清楚。在这里,我们报道了人类CIC HMG-box和C1结构域与含有一致八聚体结合位点的DNA低聚物络合的晶体结构。我们发现这两个结构域都采用了三螺旋结构,这三螺旋结构与DNA螺旋的两侧相反。C1结构域折叠成螺旋-转-螺旋(HTH)结构,插入DNA主槽以增强亲和力。我们使用分子动力学模拟和结合分析来研究该系统,这些分析询问了观察到的HMG-box和C1结构域界面以及突出的癌症变体。我们的研究结果揭示了一个独特的双部分dna结合模块,并为癌症和结构域界面突变的影响提供了见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
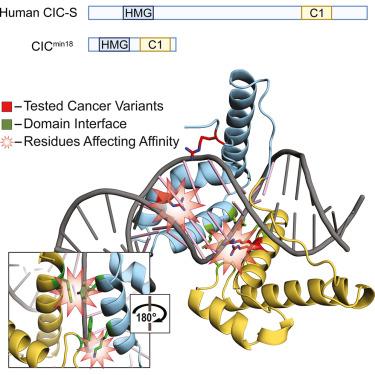
Molecular basis of DNA recognition by the HMG-box-C1 module of capicua
The HMG-box protein capicua (CIC) is a conserved transcriptional repressor with key functions in development and disease. CIC binding of DNA requires both its HMG-box and a separate domain called C1. How these domains cooperate to recognize specific DNA sequences is not known. Here, we report the crystal structure of the human CIC HMG-box and C1 domains complexed with a DNA oligomer containing a consensus octameric binding site. We find that both domains adopt tri-helical structures that pack against opposite sides of the DNA helix. The C1 domain folds into a helix-turn-helix (HTH) structure, inserting into the DNA major groove to enhance affinity. We investigate the system using molecular dynamics simulations and binding assays that interrogate the observed HMG-box and C1 domain interface and prominent cancer variants. Our results reveal a unique bipartite DNA-binding module and provide insights into the effects of cancer and domain interface mutations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


