MSnLib:高效生成开放的多级碎片质谱库。
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
非靶向高分辨率质谱是临床代谢组学、天然产物发现和暴露组学的关键工具,但化合物鉴定仍然是主要瓶颈。目前,标准工作流程应用光谱库匹配串联质谱(MS2)碎片数据。多级破碎(Multi-stage fragmentation, MSn)可以更深入地了解亚结构,从而验证破碎路径;然而,社区缺乏开放的多种天然产品和其他化学物质的MSn参考数据。本文介绍了MSnLib,这是一个机器学习准备的开放资源,包含30008个独特小分子的MSn树中的bb200万个光谱,并在开源软件mzmine中构建了高通量数据采集和处理管道。本文章由计算机程序翻译,如有差异,请以英文原文为准。
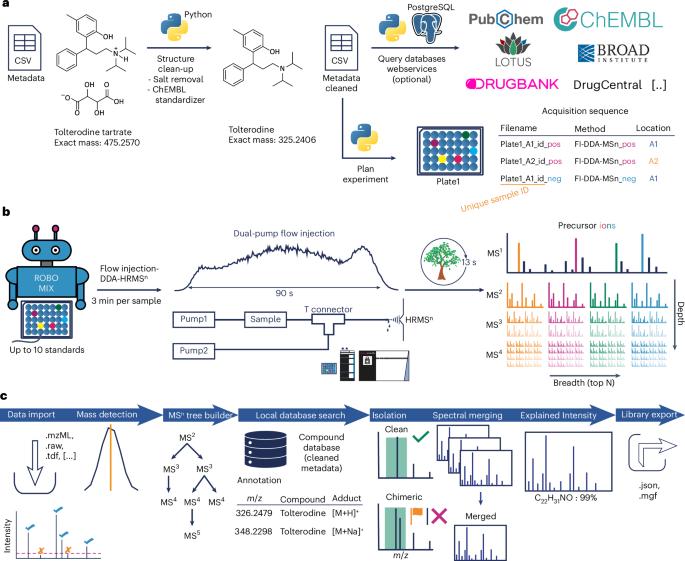
MSnLib: efficient generation of open multi-stage fragmentation mass spectral libraries
Untargeted high-resolution mass spectrometry is a key tool in clinical metabolomics, natural product discovery and exposomics, with compound identification remaining the major bottleneck. Currently, the standard workflow applies spectral library matching against tandem mass spectrometry (MS2) fragmentation data. Multi-stage fragmentation (MSn) yields more profound insights into substructures, enabling validation of fragmentation pathways; however, the community lacks open MSn reference data of diverse natural products and other chemicals. Here we describe MSnLib, a machine learning-ready open resource of >2 million spectra in MSn trees of 30,008 unique small molecules, built with a high-throughput data acquisition and processing pipeline in the open-source software mzmine. MSnLib is a large-scale, open MSn spectral library featuring >2.3 million MSn and >357,000 MS2 spectra for 30,008 unique small molecules.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


