利用深度学习和GHIST在组织学上的单细胞分辨率的空间基因表达。
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
空间分解转录组学的增加使用为疾病机制提供了新的生物学见解。然而,这些方法的高成本和复杂性阻碍了它们的广泛应用。因此,方法已经创建预测基于斑点的基因表达从常规收集的组织学图像。最近的基准测试表明,目前的方法具有有限的准确性和空间分辨率,限制了翻译能力。在这里,我们介绍了GHIST,这是一个基于深度学习的框架,通过利用亚细胞空间转录组学和多层生物信息之间的协同关系来预测单细胞分辨率的空间基因表达。我们使用公共数据集和癌症基因组图谱数据验证了GHIST,证明了它在不同空间分辨率下的灵活性和优越的性能。我们的研究结果强调了单细胞空间基因表达测量的计算机生成的实用性,以及用空间分辨组学模式丰富现有数据集的能力,为可扩展的多组学分析和生物标志物鉴定铺平了道路。本文章由计算机程序翻译,如有差异,请以英文原文为准。
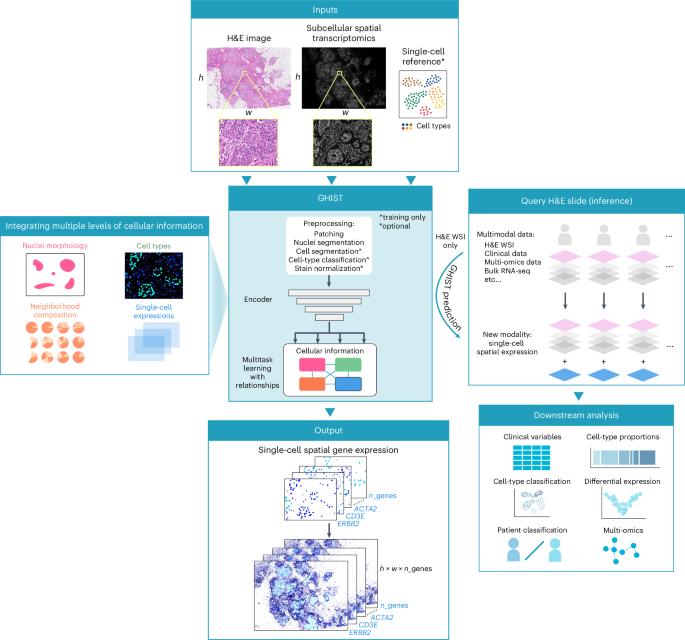
Spatial gene expression at single-cell resolution from histology using deep learning with GHIST
The increased use of spatially resolved transcriptomics provides new biological insights into disease mechanisms. However, the high cost and complexity of these methods are barriers to broader application. Consequently, methods have been created to predict spot-based gene expression from routinely collected histology images. Recent benchmarking showed that current methodologies have limited accuracy and spatial resolution, constraining translational capacity. Here, we introduce GHIST, a deep learning-based framework that predicts spatial gene expression at single-cell resolution by leveraging subcellular spatial transcriptomics and synergistic relationships between multiple layers of biological information. We validated GHIST using public datasets and The Cancer Genome Atlas data, demonstrating its flexibility across different spatial resolutions and superior performance. Our results underscore the utility of in silico generation of single-cell spatial gene expression measurements and the capacity to enrich existing datasets with a spatially resolved omics modality, paving the way for scalable multi-omics analysis and biomarker identification. GHIST is a deep learning-based method that can predict spatial gene expression at high resolution using histology data.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


