用Spacedust重新发现微生物基因组中的保守基因簇。
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
宏基因组学已经彻底改变了环境和人类相关的微生物组研究。然而,具有已知生物过程和分子功能的蛋白质的有限部分是一个主要瓶颈。在原核生物和病毒中,进化倾向于将参与相同生物过程的基因定位为保守的基因簇。相反,基因邻域的保守表明功能关联。在这里,我们提出Spacedust,一个系统的工具,从头发现保守的基因簇。为了找到同源蛋白匹配,Spacedust采用了与Foldseek快速、灵敏的结构比较。使用新的聚类和阶守恒P值检测部分保守的聚类。我们通过对1308个细菌基因组的全对比分析证明了Spacedust的敏感性,鉴定出72843个保守基因簇,其中包含420万个基因中的58%。它恢复了95%的抗病毒防御系统簇,这些簇由专用工具PADLOC注释。Spacedust的高灵敏度和高速度将有助于对大量已测序的细菌、古细菌和病毒基因组进行注释。本文章由计算机程序翻译,如有差异,请以英文原文为准。
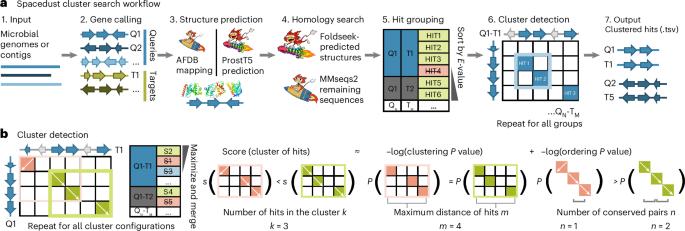
De novo discovery of conserved gene clusters in microbial genomes with Spacedust
Metagenomics has revolutionized environmental and human-associated microbiome studies. However, the limited fraction of proteins with known biological processes and molecular functions presents a major bottleneck. In prokaryotes and viruses, evolution favors keeping genes participating in the same biological processes colocalized as conserved gene clusters. Conversely, conservation of gene neighborhood indicates functional association. Here we present Spacedust, a tool for systematic, de novo discovery of conserved gene clusters. To find homologous protein matches, Spacedust uses fast and sensitive structure comparison with Foldseek. Partially conserved clusters are detected using novel clustering and order conservation P values. We demonstrate Spacedust’s sensitivity with an all-versus-all analysis of 1,308 bacterial genomes, identifying 72,843 conserved gene clusters containing 58% of the 4.2 million genes. It recovered 95% of antiviral defense system clusters annotated by the specialized tool PADLOC. Spacedust’s high sensitivity and speed will facilitate the annotation of large numbers of sequenced bacterial, archaeal and viral genomes. This work presents Spacedust, a tool for de novo identification of conserved gene clusters from metagenomic data.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


