肠道微生物群和代谢物驱动小鼠慢性镰状细胞病疼痛
IF 18.7
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
摘要
个体镰状细胞病(SCD)遭受衰弱的慢性疼痛,没有明确的病因。最近的16S核糖体RNA基因测序研究显示SCD患者存在肠道生态失调。然而,尚不清楚这些肠道微生物变化是否导致慢性SCD疼痛。使用转基因SCD小鼠,我们确定从健康对照中移植粪便微生物群后,慢性SCD疼痛得到缓解,特别是通过增加益生菌嗜粘液阿克曼氏菌的相对丰度。反过来,SCD肠道微生物组移植通过胆红素-迷走神经TRPM2信号传导诱导野生型受体持续疼痛。来自SCD患者的生物标本和人类结节神经节组织的空间转录组学分析发现了额外的细菌种类和神经元表达的转录物,应该作为新的SCD镇痛靶点进行探索。本文章由计算机程序翻译,如有差异,请以英文原文为准。
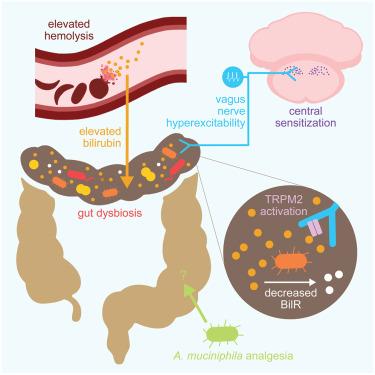
Gut microbiota and metabolites drive chronic sickle cell disease pain in mice
Individuals with sickle cell disease (SCD) suffer from debilitating chronic pain that does not have a clear etiology. Recent 16S ribosomal RNA gene sequencing studies revealed gut dysbiosis in individuals with SCD. It is unclear, however, whether these intestinal microbial changes contribute to chronic SCD pain. Using transgenic SCD mice, we determined that chronic SCD pain is alleviated following fecal microbiota transplantation from healthy controls, specifically by increasing the relative abundance of probiotic Akkermansia muciniphila. Reciprocally, transplantation of the SCD gut microbiome induced persistent pain in wild-type recipients via bilirubin-vagus nerve TRPM2 signaling. Biospecimens from individuals with SCD and spatial transcriptomic analysis of human nodose ganglia tissue identified additional bacterial species and neuronally expressed transcripts that should be explored as novel SCD analgesic targets.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


