采用16S rDNA测序和LC-MS分析帕金森病疼痛患者肠道菌群和血浆代谢物的变化
IF 2.6
4区 医学
Q3 NEUROSCIENCES
引用次数: 0
摘要
疼痛是帕金森病(PD)常见的非运动症状,严重影响患者的生活质量。近年来,肠脑轴在帕金森病发病机制中的作用引起了人们的广泛关注。然而,肠道菌群诱导的血浆代谢物变化与pd相关疼痛之间的关系仍然知之甚少。为了解决这一问题,我们分析了64名患有疼痛的PD患者(PDP), 36名患有疼痛的非PD患者(nPDP)和50名健康对照(HC)。利用16S rDNA基因测序和超高效液相色谱-串联质谱(LC-MS),我们对这些组的肠道微生物群和血浆代谢物进行了表征和比较。此外,还进行了相关分析,以确定微生物和代谢物数据之间有意义的关联。我们的发现揭示了PDP、nPDP和HC组之间的显著差异,特别是在类群中,如拟杆菌门、γ变形菌门和prausnitzii Faecalibacterium。KEGG功能预测表明细菌菌落功能主要与碳水化合物代谢、氨基酸代谢和能量代谢有关。血浆代谢组学分析发现,nPDP组和PDP组之间积累的代谢物有16种差异,其中12-酮脱氧胆酸(12-KCAc)、二氢瓦阿因(DHO)和溶血磷脂酸(LysoPA)贡献显著。LysoPA与Faecalibacterium呈显著负相关,而12-酮脱氧胆酸与Clostridium和Romboutsia呈显著正相关。该研究表明,PDP患者具有不同的肠道微生物群和代谢谱,这可能有助于PD患者的疼痛体验。这些不同的微生物和代谢物特征可能提供了将肠-脑轴与PD疼痛联系起来的潜在机制。本文章由计算机程序翻译,如有差异,请以英文原文为准。
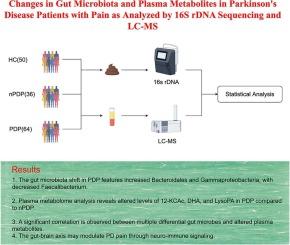
Changes in gut microbiota and plasma metabolites in Parkinson’s disease patients with pain as analyzed by 16S rDNA sequencing and LC-MS
Pain is a common non-motor symptom of Parkinson’s disease (PD) that significantly impacts patients’ quality of life. Recently, the role of the gut-brain axis in the pathogenesis of PD has garnered considerable attention. However, the relationship between gut microbiota-induced changes in plasma metabolites and PD-associated pain remains poorly understood. To address this, we analyzed 64 PD patients with pain (PDP), 36 non-PD individuals with pain (nPDP), and 50 healthy controls (HC). Using 16S rDNA gene sequencing and ultra-high-performance liquid chromatography-tandem mass spectrometry (LC-MS), we characterized and compared the gut microbiota and plasma metabolite profiles across these groups. Additionally, correlation analyses were conducted to identify meaningful associations between microbial and metabolite data. Our findings revealed significant differences among the PDP, nPDP, and HC groups, particularly in taxa such as Bacteroidales, Gammaproteobacteria, and Faecalibacterium prausnitzii. KEGG function prediction indicated that bacterial colony function was primarily related to carbohydrate metabolism, amino acid metabolism, and energy metabolism. Plasma metabolomic analysis identified 16 differentially accumulated metabolites between the nPDP and PDP groups, with notable contributions from 12-ketodeoxycholic acid (12-KCAc), dihydroouabain (DHO), and lysophosphatidic acid (LysoPA). Furthermore, a significant negative correlation was observed between LysoPA and Faecalibacterium, while 12-ketodeoxycholic acid exhibited a positive correlation with Clostridium and Romboutsia. This study suggests that PDP patients have distinct gut microbiota and metabolic profiles, which may contribute to the pain experience in PD patients. These divergent microbial and metabolite signatures may provide insight into potential mechanisms linking the gut-brain axis to pain in PD.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Brain Research
医学-神经科学
CiteScore
5.90
自引率
3.40%
发文量
268
审稿时长
47 days
期刊介绍:
An international multidisciplinary journal devoted to fundamental research in the brain sciences.
Brain Research publishes papers reporting interdisciplinary investigations of nervous system structure and function that are of general interest to the international community of neuroscientists. As is evident from the journals name, its scope is broad, ranging from cellular and molecular studies through systems neuroscience, cognition and disease. Invited reviews are also published; suggestions for and inquiries about potential reviews are welcomed.
With the appearance of the final issue of the 2011 subscription, Vol. 67/1-2 (24 June 2011), Brain Research Reviews has ceased publication as a distinct journal separate from Brain Research. Review articles accepted for Brain Research are now published in that journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


