KRAS G12突变的人肺腺癌的综合蛋白质组学特征揭示了分子发病机制
IF 13
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
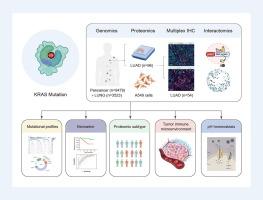
肺腺癌(LUAD)的综合蛋白质基因组学研究提供了具有潜在临床效果的独特见解。需要全面的蛋白质组学分析来更好地了解KRAS G12突变的LUAD的分子景观。目的研究KRAS G12突变LUAD的蛋白质基因组学特征。方法我们对9479例实体瘤进行了下一代测序(NGS),其中包括3523例肺癌。采用液相色谱-串联质谱(LC-MS/MS)对96例KRAS G12突变或野生型LUAD患者进行蛋白质组学分析。通过多重免疫组化(mIHC)表征LUAD肿瘤免疫微环境(TIME),并使用基于生物素的接近标记绘制KRAS g12突变肺癌细胞中的蛋白-蛋白相互作用(PPIs)。结果基因组分析揭示了KRAS在泛癌和肺癌中的异质性突变谱,并具有KRAS G12突变的等位基因特异性分辨率。KRAS g12突变体LUAD的蛋白质组学分析描绘了不同的分子特征和肿瘤进展标志。无监督聚类鉴定出三种分子亚型,其中免疫调节亚型(S2)的特征是KRAS G12C富集,具有侵袭性临床特征,免疫治疗的潜在益处最大。LUAD的免疫景观显示KRAS g12c突变肿瘤中免疫细胞浸润增加。此外,近端蛋白质组学绘制了KRAS G12突变驱动的相互作用获得的景观,离子转运体SLC4A7在G12C变异体的免疫调节中成为潜在的效应体。结论:这项全面的蛋白质基因组学研究揭示了携带KRAS G12突变的LUAD的分子特征,可能为患者分层和潜在的治疗方法提供信息,有待进一步的临床验证。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Integrative proteomic characterization of human lung adenocarcinoma with KRAS G12 mutations reveals molecular pathogenesis
Introduction
Integrated proteogenomic studies of lung adenocarcinoma (LUAD) have provided unique insights with potential clinical effects. Comprehensive proteomic analyses are needed to better understand the molecular landscape of LUAD with KRAS G12 mutations.Objectives
This study aims to characterize the proteogenomic profile of LUAD with KRAS G12 mutation variants.Methods
We performed next-generation sequencing (NGS) on 9,479 solid tumors, including 3,523 lung cancers. Proteomic profiling was conducted on 96 LUAD patients with KRAS G12 mutations or wild-type status using liquid chromatography-tandem mass spectrometry (LC-MS/MS). The LUAD tumor immune microenvironment (TIME) was characterized by multiplex immunohistochemistry (mIHC), and protein–protein interactions (PPIs) in KRAS G12-mutant lung cancer cells were mapped using biotin-based proximity labeling.Results
Genomic analysis revealed heterogeneous KRAS-informed mutational profiles across pan-cancer and lung cancer, with allele-specific resolution for KRAS G12 mutations. Proteomic profiling of KRAS G12-mutant LUAD delineated distinct molecular features and tumor progression hallmarks. Unsupervised clustering identified three molecular subtypes, with the immune modulation subtype (S2) characterized by KRAS G12C enrichment, aggressive clinical features, and the greatest potential benefit from immunotherapy. The immune landscape of LUAD showed increased immune cell infiltration in KRAS G12C-mutant tumors. Additionally, proximal proteomics mapped a landscape of gain-of-interactions driven by KRAS G12 mutations, with ion transporter SLC4A7 emerged as a potential effector in immune modulation in the G12C variant.Conclusions
This comprehensive proteogenomic study provides insights into the molecular features of LUAD harboring KRAS G12 mutations, which may inform patient stratification and potential therapeutic approaches, pending further clinical validation.求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


