寨卡病毒包膜蛋白潜在抑制剂分子对接及动力学模拟分析
IF 2.7
4区 医学
Q3 VIROLOGY
引用次数: 0
摘要
寨卡病毒(ZIKV)感染仍然是全球健康威胁,迄今为止没有批准的抗病毒药物或疫苗,因此迫切需要针对寨卡病毒的治疗方法。病毒包膜(E)蛋白对宿主细胞进入至关重要,是抗病毒干预的有效靶标。在这里,我们旨在利用双相硅筛选策略鉴定能够抑制ZIKV E蛋白的天然类黄酮化合物。首先,我们通过密度泛函理论(DFT)计算优化了9种候选类黄酮的结构,并获得了量子化学描述符(电子性质);我们还评估了他们的药物相似性和ADMET特征。其次,我们将优化后的黄酮类化合物与E蛋白进行分子对接,然后进行混合量子力学/分子力学(QM/MM)细化和100 ns分子动力学(MD)模拟,采用主成分分析(PCA)和MM- pbsa结合自由能计算来评估结合相互作用和配合物稳定性。对接发现槲皮素、匹诺曹皮素和柚皮素为最佳结合物,结合能分别为-8.3、-8.1和-8.0 kcal/mol。这些类黄酮铅还表现出良好的药代动力学特性,包括高预测胃肠道吸收、高效清除和最小毒性风险(无致癌或器官特异性警报)。值得注意的是,在100 ns MD模拟中,匹诺松蛋白复合物表现出最大的稳定性,保持紧密结合的构象。总之,槲皮素、皮诺松素和柚皮素是有前途的ZIKV E蛋白抑制剂,具有强大的靶点结合和良好的药物样谱。它们作为抗病毒候选药物的巨大翻译潜力值得进一步的体外和体内研究来证实其有效性和安全性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
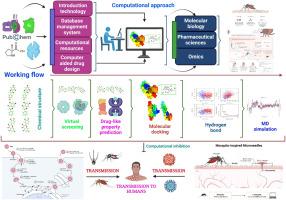
Evaluation of Potential Inhibitors of Zika Virus Envelope Protein Through Molecular Docking and Molecular Dynamics Simulation
Zika virus (ZIKV) infection remains a global health threat with no approved antivirals or vaccines to date, creating an urgent need for therapeutics targeting ZIKV. The viral envelope (E) protein is critical for host cell entry and represents a validated target for antiviral intervention. Here, we aimed to identify natural flavonoid compounds capable of inhibiting the ZIKV E protein using a dual-phase in silico screening strategy. First, we performed density functional theory (DFT) calculations to optimize the structures of nine candidate flavonoids and obtain quantum chemical descriptors (electronic properties); we also evaluated their drug-likeness and ADMET profiles. Second, we conducted molecular docking of these optimized flavonoids to the E protein, followed by hybrid quantum mechanics/molecular mechanics (QM/MM) refinement and 100 ns molecular dynamics (MD) simulations with principal component analysis (PCA) and MM-PBSA binding free energy calculations to assess binding interactions and complex stability. Docking identified quercetin, pinocembrin, and naringenin as the top binders, with binding energies of –8.3, –8.1, and –8.0 kcal/mol, respectively. These lead flavonoids also exhibited favorable pharmacokinetic properties, including high predicted gastrointestinal absorption, efficient clearance, and minimal toxicity risk (no carcinogenic or organ-specific alerts). Notably, pinocembrin’s complex demonstrated the greatest stability throughout a 100 ns MD simulation, maintaining a tightly bound conformation. In conclusion, quercetin, pinocembrin, and naringenin emerge as promising ZIKV E protein inhibitors with robust target engagement and favorable drug-like profiles. Their significant translational potential as antiviral candidates warrants further in vitro and in vivo studies to confirm efficacy and safety.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Virus research
医学-病毒学
CiteScore
9.50
自引率
2.00%
发文量
239
审稿时长
43 days
期刊介绍:
Virus Research provides a means of fast publication for original papers on fundamental research in virology. Contributions on new developments concerning virus structure, replication, pathogenesis and evolution are encouraged. These include reports describing virus morphology, the function and antigenic analysis of virus structural components, virus genome structure and expression, analysis on virus replication processes, virus evolution in connection with antiviral interventions, effects of viruses on their host cells, particularly on the immune system, and the pathogenesis of virus infections, including oncogene activation and transduction.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


