空间伪影检测提高了药物筛选实验的再现性
IF 4.1
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
可靠和可重复的药物筛选实验对药物发现和个性化医疗至关重要。我们证明了药物板的系统实验误差如何对数据的可重复性产生负面影响,并且基于板控制的传统质量控制(QC)方法无法检测到这些空间误差。为了解决这一限制,我们开发了一种与控制无关的QC方法,该方法使用归一化残差拟合误差(NRFE)来识别药物筛选实验中的系统伪像。对PRISM药物基因组学研究中100,000个重复测量结果的分析显示,nrfe标记的实验在技术重复中可重复性降低了3倍。通过将NRFE与QC方法相结合,对来自癌症药物敏感性基因组学项目的两个数据集之间的41,762对匹配的药物细胞系进行分析,我们将跨数据集相关性从0.66提高到0.76。plateQC提供了一个强大的工具集,可用于增强基础研究和转化应用的药物筛选数据的可靠性和一致性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
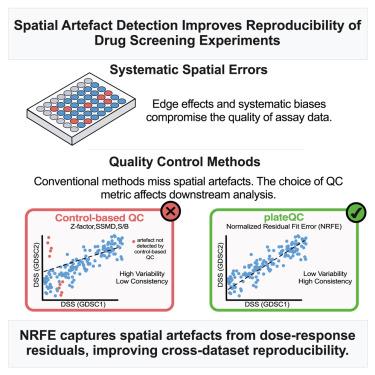
Spatial artifact detection improves the reproducibility of drug screening experiments
Reliable and reproducible drug screening experiments are essential for drug discovery and personalized medicine. We demonstrate how systematic experimental errors in drug plates negatively impact data reproducibility, and that conventional quality control (QC) methods based on plate controls fail to detect these spatial errors. To address this limitation, we developed a control-independent QC approach that uses normalized residual fit error (NRFE) to identify systematic artifacts in drug screening experiments. Analysis of >100,000 duplicate measurements from the PRISM pharmacogenomic study revealed that NRFE-flagged experiments show 3-fold lower reproducibility among technical replicates. By integrating NRFE with QC methods to analyze 41,762 matched drug-cell line pairs between two datasets from the Genomics of Drug Sensitivity in Cancer project, we improved the cross-dataset correlation from 0.66 to 0.76. Available as an R package at https://github.com/IanevskiAleksandr/plateQC, plateQC provides a robust toolset for enhancing drug screening data reliability and consistency for basic research and translational applications.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


