印度多囊卵巢综合征患者胰岛素受体胰岛素结合位点非同义snp的计算机分析及筛查
IF 0.9
Q4 GENETICS & HEREDITY
引用次数: 0
摘要
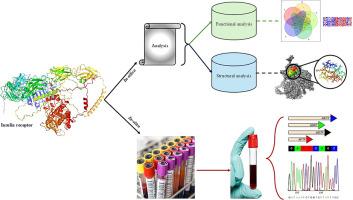
胰岛素抵抗(IR)是育龄妇女多囊卵巢综合征(PCOS)的主要病理生理特征之一。一般来说,多囊卵巢综合征的妇女与IR引起代谢综合征包括高血糖,胆固醇,和胰岛素水平由于胰岛素受体基因突变。本研究旨在利用计算机工具鉴定INSR基因胰岛素结合位点上可能存在的有害非单核苷酸多态性,并在诊断为代谢综合征的印度多囊卵巢综合征妇女中筛选这些单核苷酸多态性。从dbSNP中检索到共有7142个snp的INSR基因,其中在胰岛素结合位点发现了6个非同义snp。6个nssnp中有3个(即rs1294314902、rs1568448566、rs1353116498)与SIFT、PROVEAN、PolyPhen2、SNAP2工具预测的结果一致,而使用Project HOPE、MutPred2和MuPro工具验证了稳定性和结构变化。此外,我们采用了结构建模、对接和模拟,结果显示胰岛素配体与野生型和突变型蛋白结合的对接得分存在很大差异,这表明了nssnp的潜在危害性及其功能和结构意义。此外,我们还利用Sanger测序技术检测了63例诊断为高血糖和胰岛素抵抗的PCOS女性中INSR基因胰岛素结合位点特异性引物的snp。然而,在队列中没有发现这些nssnp。本文章由计算机程序翻译,如有差异,请以英文原文为准。
In-silico analysis of non-synonymous SNPs in insulin binding site of insulin receptor and screening in Indian PCOS women
Insulin resistance (IR) is one of the major patho-physiology of polycystic ovary syndrome (PCOS) among women of reproductive age. Generally, PCOS women with IR incur metabolic syndrome consisting of high blood glucose, cholesterol, and insulin levels due to mutations in the insulin receptor gene. Our study aimed at identifying putative deleterious nsSNPs on the insulin binding site of the INSR gene using in silico tools and screening these SNPs in the Indian PCOS women diagnosed with metabolic syndrome. The INSR gene with a total of 7142 SNPs were retrieved from dbSNP, with six non-synonymous SNPs found at the insulin-binding site. Three out of six nsSNPs (i.e. rs1294314902, rs1568448566, rs1353116498) were pathogenic as predicted using SIFT, PROVEAN, PolyPhen2, SNAP2 tools, whilst the stability and structural changes were validated using Project HOPE, MutPred2, and MuPro tools. Further, structural modelling, docking, and simulation were employed, which resulted in a wide difference between the docking scores of insulin ligand binding with wild-type and mutant proteins, indicating potential deleteriousness of the nsSNPs and their functional and structural implications. Further, SNPs were examined by utilizing Sanger sequencing using the primers specific to the insulin binding site of the INSR gene in sixty-three PCOS women diagnosed with high blood glucose and insulin resistance. However, none of these nsSNPs were found in the cohorts.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Gene Reports
Biochemistry, Genetics and Molecular Biology-Genetics
CiteScore
3.30
自引率
7.70%
发文量
246
审稿时长
49 days
期刊介绍:
Gene Reports publishes papers that focus on the regulation, expression, function and evolution of genes in all biological contexts, including all prokaryotic and eukaryotic organisms, as well as viruses. Gene Reports strives to be a very diverse journal and topics in all fields will be considered for publication. Although not limited to the following, some general topics include: DNA Organization, Replication & Evolution -Focus on genomic DNA (chromosomal organization, comparative genomics, DNA replication, DNA repair, mobile DNA, mitochondrial DNA, chloroplast DNA). Expression & Function - Focus on functional RNAs (microRNAs, tRNAs, rRNAs, mRNA splicing, alternative polyadenylation) Regulation - Focus on processes that mediate gene-read out (epigenetics, chromatin, histone code, transcription, translation, protein degradation). Cell Signaling - Focus on mechanisms that control information flow into the nucleus to control gene expression (kinase and phosphatase pathways controlled by extra-cellular ligands, Wnt, Notch, TGFbeta/BMPs, FGFs, IGFs etc.) Profiling of gene expression and genetic variation - Focus on high throughput approaches (e.g., DeepSeq, ChIP-Seq, Affymetrix microarrays, proteomics) that define gene regulatory circuitry, molecular pathways and protein/protein networks. Genetics - Focus on development in model organisms (e.g., mouse, frog, fruit fly, worm), human genetic variation, population genetics, as well as agricultural and veterinary genetics. Molecular Pathology & Regenerative Medicine - Focus on the deregulation of molecular processes in human diseases and mechanisms supporting regeneration of tissues through pluripotent or multipotent stem cells.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


