设计严格正交的生物传感器,最大限度地提高可再生生物燃料的生产过剩
IF 13
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
转录因子(transcription factors, TFs)通过结合特定的信号分子(signal molecules, SMs)来激活转录起始,然而设计能够精确靶向非天然信号分子的转录因子仍然具有挑战性。目的以转录激活子BmoR为例,建立基于机器学习的BT模型,预测三个关键残基区(crr)。本研究旨在实现严格SM正交性(SSO)的BmoR。方法采用随机森林算法建立关键残差区(CRRs)定位模型。利用Discovery Studio的计算流水线对BT模型预测数据集中的bmr - sm复合物进行了批量模拟。通过微尺度热泳(MST)亲和实验验证,对CRRs中残基进行半合理工程处理产生了具有严格SM正交性(SSO)的BmoR突变体。利用基于sso的bor生物传感器筛选3-L补料分批发酵的微生物过量生产者。结果245个TF-SM复合物的转录激活效应得到了实验验证,为生成准确率为88.5 %的机器学习模型BT提供了训练和测试数据。Discovery Studio模拟了5700个bor突变体与4个SMs之间的结合,产生了22800个配合物来输出bor - sm氢键(BSH)计数。BSH计数与补充参数结合形成预测数据集。模型BT成功预测了含有36个残基的基因突变体,并对基因突变体进行了半理性修饰,得到了带有单点登录的BmoR突变体。基于sso的bor生物传感器有效地筛选了产12.6 g/L异戊醇的菌株。结论通过证明HB在TF- sm相互作用中的主导作用,建立了TF进化的机器学习指导框架,提出了具有精确分子识别的工程TF的合理设计原则,在合成生物学和代谢工程中具有广泛的应用前景。本文章由计算机程序翻译,如有差异,请以英文原文为准。
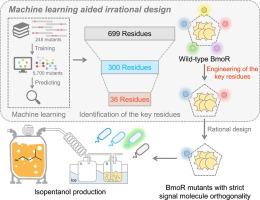
Design of strictly orthogonal biosensors for maximizing renewable biofuel overproduction
Introduction
Transcription factors (TFs) activate transcriptional initiation by binding specific signal molecules (SMs), yet designing TFs to precisely target non-natural SMs remains challenging.Objectives
Using transcriptional activator BmoR as an example, a machine-learning based model named BT to predict three crucial residue regions (CRRs) was generated. This study aimed to achieve BmoR with strict SM orthogonality (SSO).Methods
Random forest algorithm was used to generate a model BT that pinpointed crucial residue regions (CRRs). The BmoR-SM complexes in the prediction dataset of Model BT were batch-simulated using a computational pipeline via Discovery Studio. Semi-rational engineering of the residues in the CRRs generated BmoR mutants with strict SM orthogonality (SSO), validated through MicroScale Thermophoresis (MST) affinity assays. The SSO-enabled BmoR-based biosensor was used to screen microbial overproducers for 3-L fed-batch fermentation.Results
The transcription activation effects of 245 TF-SM complexes were experimentally verified, providing the training and test dataset to generate a machine-learning based model BT with 88.5 % accuracy. The binding between 5,700 BmoR mutants and four SMs was simulated by Discovery Studio, generating 22,800 complexes to output BmoR-SM hydrogen bond (BSH) counts. BSH counts combined with supplementary parameters to form a prediction dataset. The CRRs containing totally 36 residues were successfully predicted by Model BT. The CRRs were semi-rational modified to obtain BmoR mutants with SSO. The SSO-enabled BmoR-based biosensor effectively screened a strain yielding 12.6 g/L isopentanol.Conclusion
By demonstrating the dominant role of the HB in TF-SM interactions and establishing a machine learning-guided framework for TF evolution, this work advances rational design principles for engineering TFs with precise molecular recognition, offering broad applications in synthetic biology and metabolic engineering.求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


